commit 1ebf48704fd608324a690218f4d5a1b9e1054992
Author: C. Beau Hilton <cbeauhilton@gmail.com>
Date: Mon, 22 Jun 2020 09:15:31 -0500
migrating
Diffstat:
19 files changed, 1683 insertions(+), 0 deletions(-)
diff --git a/2018-09-01-on-making-this-website.md b/2018-09-01-on-making-this-website.md
@@ -0,0 +1,164 @@
+---
+layout: post
+title: "On the tools used to make this website"
+toc: false
+categories:
+ - technical
+tags:
+ - jekyll
+ - markdown
+ - atom
+ - hosting
+ - static web
+ - blog
+ - technical
+---
+
+I'm psyched about this little website. This post is about the tools and resources I used to make it.
+
+In summary, this is a _static website_ made with _Jekyll software_ using the code editor called _Atom_ to write posts in _Markdown language_. I also added a wishlist to the end, mostly to keep track of tools and tricks I may want to add.
+
+## Static Web
+
+[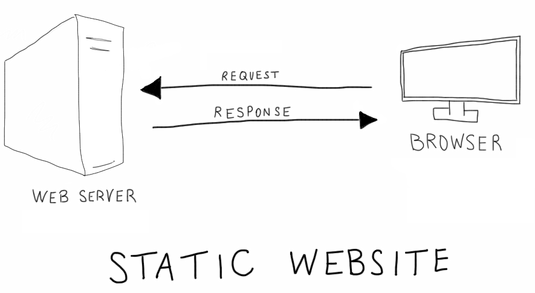](https://blog.zipboard.co/how-to-start-with-static-sites-807b8ddfecc)
+
+Most of the internet we interact with on a day-to-day basis is "dynamic," meaning we request something from a web server (e.g. click on a Facebook post or search for something on Amazon), which then talks to a database to pull all the relevant information and fill in a template (e.g. post content + comments, or product page with images, ratings, ads, etc.). The "static" web is much more old school: ask for something and the server gives you something, no database and comparatively little processing. Whatever is on the server has to be pretty much prebuilt and ready to go. This process is much simpler, faster, and more secure. The basic protocols are old and well established, unlikely to change or break. For most online writing, such as a blog, static is the way to go, especially if it is likely to be consumed over a mobile internet connection.
+
+## Jekyll
+
+[](https://jekyllrb.com/)
+
+Jekyll is an open-source suite of open-source software for making static web pages.
+
+The basic idea is create a workflow that starts with just writing, followed by a predefined file structure and styling format to store all your stuff and make it pretty, and finally automatically translate it all into something a web browser can use. Jekyll also integrates nicely with GitHub, [which will host the site for free](https://help.github.com/articles/about-github-pages-and-jekyll/). GitHub also makes my favorite text editor, [Atom](https://atom.io/). This leads to an integrated workflow: write in Atom → build in Jekyll+GitHub → publish on GitHub.
+
+I found out about Jekyll from the [Rebecca Stone](https://ysbecca.github.io/programming/2018/05/22/py-wsi.html), who is writing the wonderful [py-wsi package](https://github.com/ysbecca/py-wsi). (This package makes massive digital pathology images tractable for deep learning.) As I read through her blog, I found [this post on Jekyll and why she switched from WordPress](https://ysbecca.github.io/programming/2017/04/29/jekyll-migration.html). I read a few more posts like hers and was converted.
+
+Some user-friendly Jekyll walkthroughs:
+
+- [Programming Historian: complete Jekyll and GitHub pages walkthrough for non-programmers](https://programminghistorian.org/en/lessons/building-static-sites-with-jekyll-github-pages)
+ - Assumes you know absolutely nothing, explains terminology and has copy-paste code plus instructions for Windows *and* Mac.
+- [WebJeda: YouTube videos](https://www.youtube.com/watch?v=bwThn0rxv7M)
+ - Modular and step-by-step. Sometimes it's nice to watch someone go through the process in real time.
+- [University of Idaho Library: workshop, with blog posts and video](https://evanwill.github.io/go-go-ghpages/0-prep.html)
+ - Comparable to Programming Historian, with the benefit of dual coverage in the blog and video. The video is from a workshop where they walked through the process in real time, from nothing to something in around an hour, and the students asked a lot of the same questions I had.
+
+### Jekyll Themes
+
+I stole the layout of this site from [vangeltzo.com](https://vangeltzo.com/index.html), whose beautiful design was Jekyll-ified (with permission!) by [TaylanTatli on GitHub](https://taylantatli.github.io/Halve/). I'm [not the only one](https://github.com/cbeauhilton/cbeauhilton.github.io/network/members) using this theme, [by](https://drivenbyentropy.github.io/) [any](https://ejieum.github.io/) [means](https://je553.github.io/).
+
+Jekyll themes are [abundant online](http://jekyllthemes.org/), generally [easy to fork](https://taylantatli.github.io/Halve/halve-theme/) ("fork" means "copy for your own use without affecting the original"), and often [well commented](https://taylantatli.github.io/Halve/posts) so they are straightforward to customize.
+
+### Why this theme?
+
+I had two basic requirements for my theme. First, it had to be [pretty](http://www.leonardkoren.com/lkwh.html). Second, I wanted a responsive split screen. "Responsive" means it can adapt itself to look nice on any device from a phone to a huge desktop monitor. The split screen confines the content to an easily readable width on the right and gives consistent navigation on the left. The 50/50 split screen is a little extreme, but I think I like it this way. If I ever want to change it to something like 66/33, it is [quite easy](https://github.com/TaylanTatli/Halve/issues/32).
+
+I'll say more about specific choices, such as color, logo, and tagline in another post.
+
+## Atom
+
+[](https://atom.io/)
+
+You can write all of your code entirely within GitHub, which has its own text editor. The above tutorials use this approach to make it easy to start. There's nothing wrong with that.
+
+A friend of mine, a [fellow CCLCM student](https://github.com/JaretK) who is a much more legit programmer than me, recommended Atom, and I fell in love with only minimal doses of digital [amortentia](http://harrypotter.wikia.com/wiki/Amortentia). It is ridiculously (almost dangerously) extensible, with community-supplied packages for an incredible breadth of programming languages and applications.
+
+For the nerds, for example, it has full support for LaTeX and all the typesetting, figure-making, citation-porn, and mathy goodness you can handle. One of my favorite workflows is to write something in Markdown to get the basics down with minimal fuss in an easier language, and then use Pandoc to convert it to LaTeX, followed by any futzing with the LaTeX (if you like) before outputting to PDF. You can also (!) combine the languages rather seamlessly.
+
+Here's a [post that details a full plain-text academic workflow with Atom](http://u.arizona.edu/~selisker/post/workflow/).
+
+For the poets, it can be a gorgeous and focused writing environment with infinitely more flexibility than Word. Here's [a post that describes how one creative writer uses Atom](https://8bitbuddhism.com/2017/12/29/a-novel-approach-to-writing-with-atom-and-markdown/). The [Zen package](https://atom.io/packages/Zen) alone should entice you Hemingway types, but also take a look at the writing assistance tools detailed in previous hyperlink or the hyperlink in the next paragraph.
+
+For me, the coolest thing is that I can have [one central hub](https://medium.com/@sroberts/how-i-atom-12988bce8fce) to do it all, with very little need for Word. If I use Atom to push things to GitHub, it also nearly replaces Google Docs for keeping everything safely in the cloud (caveat for the latter:as long as I don't mind making my work public).
+
+Note from 2019: I switched to [VS Code](https://code.visualstudio.com/). It's very similar to Atom, but faster.
+
+## Markdown
+[](https://daringfireball.net/projects/markdown/)
+
+Markdown is a "text-to-HTML conversion tool for web writers [that] allows you to write using an easy-to-read, easy-to-write plain text format, then convert it to structurally valid XHTML (or HTML)." (click image or [here](https://daringfireball.net/projects/markdown/) for source).
+
+It's inspired by the way people used to mark up their plain-text emails to make them more readable, with all the "#" and "-" and "***" you can handle. Therefore, if you look at the source of something written in Markdown, it's still pretty darn legible. Those of you who have read and written medical charts, many of these are still in common use (the "#" problem list, for example).
+
+Here are examples from a great [GitHub Markdown cheatsheet](https://github.com/adam-p/markdown-here/wiki/Markdown-Cheatsheet). First is the code and then the rendered text.
+
+`Emphasis, aka italics, with *asterisks* or _underscores_.`
+Emphasis, aka italics, with *asterisks* or _underscores_.
+
+`Strong emphasis, aka bold, with **asterisks** or __underscores__.`
+Strong emphasis, aka bold, with **asterisks** or __underscores__.
+
+`Combined emphasis with **asterisks and _underscores_**.`
+Combined emphasis with **asterisks and _underscores_**.
+
+```
+Section with a hashtag:
+# H1
+```
+
+Section with a hashtag:
+# H1
+
+
+```
+Section using underline style to accomplish the same thing:
+Alt-H1
+======
+```
+
+Section using underline style to accomplish the same thing:
+
+Alt-H1
+======
+
+
+Pretty cool, eh?
+
+## OK, so what do you _actually_ do?
+
+- Install all the stuff from one of the Jekyll how-tos mentioned above, follow one of the guides to set up GitHub pages, Travis CI, and your local jekyll environment.
+- Open a new `.md` file in your favorite text editor.
+- Insert something like this at the top:
+
+```
+---
+layout: post
+title: "On the tools used to make this website"
+toc: false #table of contents
+categories:
+ - technical
+tags:
+ - jekyll
+ - markdown
+ - atom
+ - hosting
+ - static web
+ - blog
+ - technical
+---
+
+```
+
+- Write all your stuff using Markdown syntax.
+- Commit your changes to GitHub
+- Wait for Travis CI to build, fix any errors (for example, you might need to delete the `BUNDLED WITH` lines in the gemfile.lock).
+- Check out your new post!
+
+## Wishlist/possibilities
+
+
+- Clickable Zen *reading* mode (remove all hyperlinks - whether or not hyperlinks impair focus is [hotly debated](https://books.google.com/books?id=QJxeBAAAQBAJ&pg=PA79&lpg=PA79&dq=On+Measuring+the+Impact+of+Hyperlinks+on+Reading&source=bl&ots=Ih_zN17-Nh&sig=F47u2HB7nBavnD3amydmJo5wNB4&hl=en&sa=X&ved=2ahUKEwjs0Pif1Z3dAhUKXa0KHcEeCucQ6AEwCXoECAEQAQ#v=onepage&q&f=false), he said with a meta-smirk).
+- Clickable dark/light theme switch, a la [dactl](https://melangue.github.io/dactl//) (that black and white water drop button on the top right) or [Hagura](https://blog.webjeda.com/dark-theme-switch/) (the text that reads "Dark/Light" at the bottom of the page is a button), for kinder day and night reading. The Hagura author wrote a tutorial [here](https://blog.webjeda.com/dark-theme-switch/).
+- ~~Side notes, a la Tufte. [Michael Nielsen's blog](http://augmentingcognition.com/ltm.html) has one implementation, and this [Tufte-ite Jekyll theme](http://clayh53.github.io/tufte-jekyll/articles/15/tufte-style-jekyll-blog) has another. I would probably have to reduce the page split from 50/50 to at least 66/33 for this to work properly.~~
+- ~~Or: [Barefoot footnotes](https://github.com/philgruneich/barefoot) (click and they show up, like Wikipedia)~~ Barefoot works very well, check out other posts.
+- Integrate fancy plugins
+
+ - ~~[Overview of how to integrate non GitHub-approved plugins, with nice script that automates the integration](https://drewsilcock.co.uk/custom-jekyll-plugins).~~ Went with Travis CI instead, using [these instructions](http://joshfrankel.me/blog/deploying-a-jekyll-blog-to-github-pages-with-custom-plugins-and-travisci/). I ended up breaking everything and fighting with it for a day, but it seems the whole issue was that the backend process of migrating from the github.io address to beauhilton.com was not instantaneous, and as soon as it was done I could change the internal reference to <https://beauhilton.com> and it came together. All that heroic hacking, for nothing...
+ - [Academicons](https://www.janknappe.com/blog/Integrating-Academicons-with-Fontawesome-in-the-Millennial-Jekyll-template/).
+ - [Jekyll Scholar](https://gist.github.com/roachhd/ed8da4786ba79dfc4d91) and [Jekyll Scholar Extras](https://github.com/jgoodall/jekyll-scholar-extras), see [example with clickable BibTex and PDF downloads](https://caesr.uwaterloo.ca//publications/index.html) in publications page that makes me salivate.
+ - [Integrate Jupyter Notebooks seamlessly](https://bethallchurch.github.io/jupyter-notebooks-with-jekyll/) (the way people usually do it, which is probably fine and doesn't require moar code, is to write up a plain-language explanation for the blog and link to the ipynb file on GitHub).
+ - [Add estimated reading time to pages](https://github.com/bdesham/reading_time).
+ - [Add search bar](http://www.jekyll-plugins.com/plugins/simple-jekyll-search).
+ - [Make the site pictures responsive](https://github.com/robwierzbowski/jekyll-picture-tag).
+ - [Add static comments to posts that would benefit from community](https://mademistakes.com/articles/jekyll-static-comments/#static-comments).
+ - ~~Get Table of Contents working for lengthy posts such as this one.~~ Added this feature, though not in this particular post because the "# H1" Markdown examples screw it up.
diff --git a/2018-09-07-on-the-slogan.md b/2018-09-07-on-the-slogan.md
@@ -0,0 +1,87 @@
+---
+layout: post
+title: "On the pretentious Latin slogan"
+categories:
+ - medicine
+tags:
+ - history
+ - balance
+ - humility
+ - medicine
+---
+
+# sola dosis facit venenum
+
+### Or: the dose makes the poison
+
+<p></p>
+___________
+<p></p>
+
+First, two quotes, one in English and one in German.
+
+Don't worry, we'll translate the German (this isn't one of those 19th century novels that expects fluency in English, French, Latin, Italian, and German just to get through a page), and the same basic idea is found in the English quote. In German one of the words has a delightful history that is exposed when juxtaposed with an English translation.
+
+> Poisons in small doses are the best medicines; and the best medicines in too large doses are poisonous.
+
+> <cite><a href="http://theoncologist.alphamedpress.org/content/6/suppl_2/1.long">William Withering, 18th century English physician, discoverer of digitalis (sort of), and proponent of arsenic therapy.</a></cite> [^1] [^2]
+
+>Alle Dinge sind Gift, und nichts ist ohne Gift; allein die dosis machts, daß ein Ding kein Gift sei.
+
+> <cite><a href="http://www.zeno.org/Philosophie/M/Paracelsus/Septem+Defensiones/Die+dritte+Defension+wegen+des+Schreibens+der+neuen+Rezepte"> 1538, *Septem Defensiones*, by Swiss physician Philippus Aureolus Theophrastus Bombastus von Hohenheim, also known as Paracelsus, "Father of Toxicology."</a></cite>
+
+The phrase "sola dosis facit venenum," (usually rendered "the dose makes the poison" in English) is a Latinization of the German phrase above from Paracelsus. He wrote this in his "Seven Defenses" when he was fighting against accusations of poisoning his patients[^4] (malpractice court, it seems, is one of the oldest traditions in medicine).
+
+Here's my rough translation from Paracelsus' German:
+
+> All things are poison, and nothing is not a poison. Only the dose makes the thing not a poison.
+
+Throw in a couple of exclamation marks, italics, and fist pounds, and you have the makings of dialogue for the defendent in a 16th century *Law and Order*.
+
+### Paracelsus
+
+Why did this German guy have a Latin name?
+
+No one really knows. Von Hohenheim's relationship with Latin and the humanistic antiquity it represented was complicated. Despite writing books with names such as *Septem Defensiones*, he was known for refusing to lecture in Latin, preferring vernacular German instead, and the content of his books was invariably German.[^5] He publicly burned old Latin medical texts, along with their outdated ideas. My favorite version of the Paracelsus story is that the Latin name was first given to von Hohenheim by his friends, who were probably screwing with him, as they knew it would go against his iconoclastic bent. He used it (with a glint in his eye) as an occasional pen name, and it eventually stuck.
+
+In this spirit I decided on the Latin slogan. It's a bit of a post-post-modern jab at myself, medical tradition, and old bearded white men, who sometimes knew how to make fun of themselves and occasionally had good ideas despite their bearded whiteness.[^6] Also, Latin is pretty. And regardless of language, the idea this phrase represents is possibly my favorite, in medicine and in life.
+
+### Poison and other gifts
+
+The word for "poison" in German is _Gift_. English "gift," meaning "present," comes from the same Proto-Indo-European root: *ghebh*-, "[to give](https://www.etymonline.com/word/gift)." (Language is, after all, susceptible to [divergent evolution](https://en.wikipedia.org/wiki/Divergent_evolution), sometimes [amusingly](http://www.bbc.co.uk/languages/yoursay/false_friends/german/be_careful__its_a_gift_englishgerman.shtml)). This dual meaning fits nicely with the general conceit --- medicine and poison are two sides of the same coin, gifts in either case, to be used [with judgment, not to excess](https://www.lds.org/scriptures/dc-testament/dc/59.20).
+
+We can keep going down the etymologic rabbit hole: [poison](https://www.etymonline.com/word/poison) comes from Proto-Indo-European _po(i)_, "to drink," the same root that gave us "potion" and "potable." The Latin [_venenum_](https://www.etymonline.com/word/venom) from our quote, which eventually led to English "venom," comes from Proto-Indo-European _wen_, "to desire, strive for," which made its way around to _wenes-no_, "love potion," and then to more general meaning as "drug, medical potion," before finally assuming the modern meaning of a dangerous substance from an animal. I wonder if the evolution of these words from neutral or positive to negative reflects cycles of hope and disillusionment. The history of medicine is filled with well-meaning, careful clinicians who were often simply wrong, as well as charlatans ([medicasters](https://www.etymonline.com/word/medicaster), even).
+
+By contrast, the word "medicine" has undergone remarkably little change in meaning. It comes from Proto-Indo-European [_med_](https://www.etymonline.com/word/*med-), "to take appropriate measures," a root shared with "meditate," "modest," "accomodate," and, of course, "[commode](https://www.etymonline.com/word/commode)." This is a complete way to think about how to practice and receive medicine: to take appropriate measures. The difficult part is deciding what "appropriate" means.
+
+### So what?
+
+I'm going to be a doctor shockingly soon (2020, two years after this writing). After residency I will most likely specialize in either hematology-oncology (blood cancer) or palliative care,[^3] and in either case will help people decide which poison to take in an effort to feel better.
+
+Crazily enough, some of the best treatments for some of the worst diseases are classical poisons, such as arsenic for some leukemias. Arsenic is what the lucky ones get to take --- we have and regularly use other drugs that are much more dangerous. It's not just drugs we poison people with, either. If you are fortunate enough to have the right kind of cancer, enough physical strength left in reserve, and a donor to make you eligible for a bone marrow transplant, get ready to become friends with a [linear particle accelerator](https://en.wikipedia.org/wiki/Linear_particle_accelerator). We radiate the bone marrow until it gives up the ghost, then start over fresh, resurrection from the inside out.
+
+In palliative care it's not so different, even though the goal is symptom management and not cure. An illustrative anecdote: during a palliative care rotation I was trying to help the physician do some calculations she had been doing by hand. I thought, "there has to be an online calculator for this...", and pulled up several medical calculators from respected organizations. In every case, after I put in the drug and dose I wanted, the website would say something like,
+
+>"Hell no. We don't touch that stuff. We're not even going to do the calculation for you. If you need that medicine at that dose, call a palliative care doc. Consider our asses covered."
+
+We dispense derivatives of poppy that make heroin look tame, administer amphetamines as antidepressants, and [we're starting to get good at using LSD](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5867510/). Poisons, all. Medicines, all.
+
+My smartass response when people ask me why I chose medicine is "power, drugs, money, and women." It's not entirely untrue: nobody else is allowed to use these tools on humans, whether fancy knives or fancy x-rays or fancy poisons; I will always have a job and a comfortable living no matter what happens to the economy (my wife and I have had kids since undergrad, which didn't change my career goals but added an extra layer of responsibility); and chicks dig docs in uniform (the coat is white because it's hot).
+
+### To sum up:
+
+In the right hands, with respect and care, even the most dangerous substances can be tools for healing. I love that. So, for now at least, the pretentious phrase stays. Once more, with gusto: _Sola dosis facit venenum._[^7]
+
+[^2]: The article from The Oncologist that gave us the Withering quote made the unfortunate mistake of saying he was a **15th** century physician, which has, even more unfortunately, led to the persistence of the wrong century attached to his name in other publications. He was born in 1741, discovered digitalis in 1775, and died in 1799. <p> </p> Because I'm completely insufferable: 
+
+[^1]: Foxglove, the common name for *Digitalis purpura*, has been used in medicine for centuries, most notably for "dropsy," the edema (fluid accumulation) associated with heart failure and other conditions. <p> Withering was the one who figured out extraction methods, dosages, and side effects, and most thoroughly estabilshed it as a medicine rather than a folk remedy or poison, but it didn't come out of nowhere. This is almost always the case: we stand on the shoulders of giants, aspirin comes from bark, etc. </p> <p> While I kind of wish the story that Withering stole it wholecloth from a local herbalist, Mother Hutton, was true, as it would fit my bias that most medicine is built on masculine appropriation of traditionally feminine arts without giving proper credit, this particular story [does not appear to have any historical basis](https://doi.org/10.1016/S0735-1097(85)80457-5). (by the way: amniotomy hooks are sharpened crochet hooks, surgical technique is mostly translated sewing technique, pharmacy is fancy herbalism --- and it's not just in medicine: is a chef a man or a woman? What about a cook? Men make art, women make crafts. Etc., ad nauseum.)
+
+[^3]: [Palliative care](https://getpalliativecare.org/whatis/) is all about helping people feel better when their disease or the treatment for it makes them feel crappy, physically and emotionally. Palliative care providers also know the legal system and end-of-life decision-making inside and out, and much of their work is counseling with worried and blindsided families who are trying to make decisions with and for their critically ill loved one.
+
+[^4]: If we put on our 21st century lenses, we see that he was indeed poisoning his patients, but in good faith rather than out of quackery or malice. Paracelsus himself [probably died from mercury intoxication](http://www.paracelsus.uzh.ch/general/paracelsus_life.html), incurred from years of practicing alchemy, which was a perfectly respectable occupation for the learned of the time.
+
+[^5]: One of the richest records of Paracelsus' thought is _The Basle Lectures_, which is written in Latin, but this is a [collection of class notes](http://www.paracelsus.uzh.ch/general/paracelsus_works.html) taken by his students, who, like all good med students of the time, studied in Latin. The rest of the best work from and on Paracelsus [is in German](http://www.paracelsus.uzh.ch/texts/paracelsus_reading.html), as far as I can tell, with [few exceptions](https://yalebooks.yale.edu/book/9780300139112/paracelsus). This is a source of irritation for this monolingual Anglophone.
+
+[^6]: Ok, Paracelsus wasn't bearded. But he should have been. I mean, look. 
+
+[^7]: On the pronunciation: if you want to sound more Italian, then "soh-la dose-ees fa-cheet ven-ay-noom." If you want to have fun in a different way, make the "c" in "facit" a hard "k" and throw in a comma and some emphasis: "soh-la dos-ees, **fa-keet** ven-ay-noom." Go ahead. Try it out loud.
diff --git a/2018-10-20-a-workflow-for-remembering-all-that-science-and-also-everything.md b/2018-10-20-a-workflow-for-remembering-all-that-science-and-also-everything.md
@@ -0,0 +1,107 @@
+---
+layout: post
+title: "Remember all the things"
+categories:
+ - learning
+tags:
+ - spaced repetition
+ - desirable difficulty
+ - education
+ - learning
+ - how-to
+---
+
+# We spend so much time reading
+## and forgetting.
+
+Like most of you, I read a silly number of books, journal articles, blog posts, whatever. I forget the vast majority of what I read, almost immediately. Chances are, so do you. We're humans.
+
+The brain is a conservative organ, ruthlessly efficient. It kills memories left and right unless it has a really good reason not to.
+
+The upper limit of short term memory is 6-8 items, and most of what we learn is dumped within a day.[^12] However, there is no known upper limit on long-term memory. Let that sink in: _no known upper limit_. That is, memory is **_in-finite_**: we don't know how much we can know. We are still exploring the far reaches of long-term memory, and I'm excited. What will be possible after decades of purposeful retention of _all the interesting and useful things_?[^1]
+
+Here is a system cobbled together, with the [help of many friends](https://www.mdedge.com/ccjm/article/110825/practice-management/information-management-clinicians), to convince the brain to have a little mercy.[^10] As of October 2018, I've been doing this for a few months, and it's wonderful. It is [just cumbersome enough](https://en.wikipedia.org/wiki/Desirable_difficulty) to create conditions for learning, but not so much that I find excuses to skip the process.[^13] One difference between this system and what I used to do is the proposed longevity: instead of learning things to pass a class, or do a thing, my goal is to have a coherent way to gain and keep knowledge for life, in all its domains.[^11]
+
+In order to keep this page pretty and focus on process rather than tools, I put specific technologies and commentaries in the clickable footnotes.
+
+### Find the thing:
+- discover [^2]
+- decide (is it worth my time?) [^3]
+
+### Hoard:
+- so it's available forevermore (for citation, review, sharing) [^9]
+
+### Read:
+- annotate (clarify, connect, question) [^4]
+- highlight (_only_ what I might want to _memorize_. Nothing else.) [^5]
+
+### Review:
+- scan annotations and highlights. [^6]
+- decide what to commit to long-term memory. [^7]
+
+### Retain:
+- spaced repetition with digital notecards (key points, actionable facts, so what?, lovely quotes, etc.) [^8]
+
+If you have suggestions for improvement, questions, tools, or experiences, I would love to hear them.
+
+
+[^1]: I have a friend who has kept up on his spaced repetition tools from the first US medical licensing exam (10-15k digital flashcards, all the things you learn in the first two years of medical school), and I positively seethe with envy at how little review he will have to do when he takes the second exam.
+
+ Much, if not _most_ of my preparation time for the second exam was re-learning things I had learned for the first exam and forgotten. My proof of this was that when I missed practice questions, I could usually search my old flashcard deck and find the exact answer. How much better would life be if I could keep a larger proportion of what I learn, especially if I do it with a system that takes minutes per day?
+
+ Of course, not everything needs to be retained, and the medical licensing exams are, unfortunately, riddled with trivia (i.e. things that are highly testable, but of little use clinically, such as the chromosomal location of a very rare genetic disorder: most doctors will never see the disease, and if they do the last thing that will matter is the location on the chromosome).
+
+[^2]: In order, these are my preferred ways to find things:
+ 1. **word of mouth** (in-person, ideally, but a well-crafted newsletter is also a delight. Despite all this technology, I find the best way to address the problem of "[unknown unknowns](https://en.wikipedia.org/wiki/There_are_known_knowns)" is through respected friends and colleagues).
+ 2. **purposeful search** (goal-driven: help this patient, write this paper/blog post, scratch this itch).
+ 3. **automatic digests** ([RSS feeds](https://fraserlab.com/2013/09/28/The-Fraser-Lab-method-of-following-the-scientific-literature/) of keywords, journals, authors, etc.) I find this approach less useful for students, who have usually not yet differentiated into their specific interests, because any feeds they set up will be too noisy. The link above is great for an overview and examples, but their intake funnel is too wide for my taste. For example, they include the general feed from _Nature_ in addition to highly idiosyncratic feeds. _Nature_ has some interesting stuff and is a blast to browse, but my goal with RSS is to be targeted and _save_ time: keep the idiosyncracies and ditch _Nature_, unless you set up a filter for only the types of articles from _Nature_ you actually care about.
+
+[^3]: Quick scan, check out the pictures, skim abstract if available, consider strength of recommendation. Certain people have given me such delicious recommendations in the past that anything they now recommend skips to the head of the reading list.
+
+[^4]: For most web-first content, [Diigo](https://www.diigo.com) is great, and I use [Xodo](https://www.xodo.com/) for PDF markup.
+
+ Xodo is the best free full-featured PDF tool I have found: quick, reliable, support for search, annotation (and signatures), highlighting.
+
+ [SumatraPDF](https://www.sumatrapdfreader.org/free-pdf-reader.html) is the fastest PDF reader I have found, great for pure ctrl-F search especially with huge PDF textbooks, but achieves this by being absolutely bare-bones (no handwritten annotation, highlighting, etc.).
+
+ I also love paper and pen, and because I put the things I want to keep into spaced repetition software, I can usually recycle the paper without fear of losing something important.
+
+[^5]: Highlighting is _not_ a good method to engage with text and make it sink in. This has been studied, [repeatedly](https://eric.ed.gov/?id=EJ1021069). One of the studies cited in the linked review showed that highlighting _impairs_ comprehension.
+
+ However, if the highlight signals something meaningful and consistent, such as only the kind of information you might want to memorize, you can use the visual cue to quickly find, review, and make decisions about your next steps.
+
+ I still do most of my reading on paper, where highlighting makes review easy. In an app like [Diigo](https://www.diigo.com) or Kindle, your highlights are extracted into a separate document, which tightens review even further.
+
+[^6]: Here's where highlighting only the things you might want to memorize shows its utility.
+
+ You may also find that your annotations need annotations, e.g. a question you asked was answered later in the paper.
+
+[^7]: If something seems worth the [5-10 minutes](http://augmentingcognition.com/ltm.html) it will take to make and review a flash card, do it. If not, screw it. Also, develop a low threshold for fixing or deleting cards. This makes card creation in the first place less of a daunting task.
+
+[^8]: If you keep all your digital notecards in a tool like [Anki](http://ankiguide.com/getting-started-with-anki-srs/), you will have the spaced repetition part automated, provided you remember to open the app every day.
+
+ If you keep everything in a single Anki deck, regardless of subject, you will also gain the benefit of [interleaving](https://www.scientificamerican.com/article/the-interleaving-effect-mixing-it-up-boosts-learning/). Interleaving is one of a handful of techniques shown to improve memory and understanding, and basically involves mixing material freely without regard for category.
+
+ For example, instead of homework with all quadratic equations, mix in all the math from the whole semester in random assortment. In medicine, interleaving is particularly important: if a person comes in with chest pain, heart attack is not the only possibility, and if I had studied only cardiology that month I might fail to consider panic attacks, lung problems, digestive issues, or even blunt trauma, and send the person home after a normal EKG without even looking at their chest or asking any questions. Real life is interleaved.
+
+ So I keep my Anki cards all mixed up, such that in a single session I might see bits of useful computer code, friends' birthdays, dosing regimens for drugs, key points of journal articles, beautiful quotes, etc., in rapid succession.
+
+[^9]: [Diigo](https://www.diigo.com) for standard web fare, [Zotero](https://academicguides.waldenu.edu/library/zotero) for journal articles with PDFs.
+
+ We have free, infinitely large Google Drives (for life!) at my university, so I use the [process described here](https://docs.google.com/document/d/1dmdLyZut4rpfPDF8Mt_JhgHLtYkmYS6kggkwn1lSGQQ/edit?usp=sharing) to keep the PDFs available in the cloud. If I use this I can read and markup on my phone, computer, whatever, and it will automatically sync.
+
+[^10]: This did not come out of a vacuum. I started with spaced repetition in undergrad under the guidance of a brilliant tutor, then learned from my own students, and am continually indebted to countless classmates and generous souls online.
+
+ I am deeply grateful to [Harvard Macy Institute](https://www.harvardmacy.org/index.php) and [Dr. Neil Mehta](https://www.linkedin.com/in/neilbmehta/), in particular, for pushing me to formalize my thinking and get beyond banal "cram and regurgitate" methods for exams and classrooms. They were also the first to give me a framework, and provided many of these tools.
+
+ These ideas are not new, and have been presented in many forms: my contribution here is a personal system for putting it all together, with my preferred tools. **The key addition is spaced repetition**, which makes it much more likely for ideas to be imprinted deeply and available quickly.
+
+[^11]: For the philosophic among you: I study artificial intelligence, tools that can consider trillions of variables all at once. The goal is to outsource certain types of thinking that computers are better at, for the benefit of (in my case) people with serious illness.
+
+ This fits with the basic story of technology: outsource a thing to amplify it (I can pick up a rock: a crane can pick up a house; I can run: an airplane can fly; I can remember some things: Google remembers all the things).
+
+ A few technologies, however, are about _insourcing_, that is, developing the human element to its maximum. Spaced repetition is one of these technologies, optimized with proven algorithms to maximize how much we can learn and retain. What will be possible when highly developed human memories couple with human creativity, as well as technological support from AI and other advances? So far the research on AI suggests that the very best systems are not solely computers or solely humans, but hybrids, humans working with machines, emphasizing the strengths of both. What happens when we optimize each component? For good and for bad?
+
+[^12]: The average working memory of primates, including humans, is 4 items. The range is 1-8, ish, depending on the details of the study and participants. See [EK Miller's lab page](http://ekmillerlab.mit.edu/publications/) for links to recent work, especially **Miller, E.K. and Buschman, T.J. (2015) Working memory capacity: Limits on the bandwidth of cognition. Daedalus, Vol. 144, No. 1, Pages 112-122.** A free PDF of this paper is available on the linked website.
+
+[^13]: Free PDF article on desirable difficulty from UCLA available [here](https://bjorklab.psych.ucla.edu/wp-content/uploads/sites/13/2016/04/EBjork_RBjork_2011.pdf).
diff --git a/2018-11-01-Artificial-Intelligence-Definitions-and-Indefinitions.md b/2018-11-01-Artificial-Intelligence-Definitions-and-Indefinitions.md
@@ -0,0 +1,132 @@
+---
+layout: post
+title: "On the buzziest of buzzwords: what is AI, anyway?"
+toc: true
+image: /images/unsplash-grey-flowerbuds.jpg
+categories:
+ - AI for MDs
+tags:
+ - artificial intelligence
+ - machine learning
+ - deep learning
+ - big data
+ - definitions
+---
+
+# Introduction
+
+
+
+Artificial Intelligence is kind of a big deal.[^2] Despite real advances, particularly in medicine, for most clinicians "AI" is at best a shadowy figure, a vaguely defined ethereal mass of bits and bytes that lives in Silicon Valley basements and NYT headlines.
+
+"Bob, the AI is getting hungry."
+
+"I don't know, Jane, just throw some AI on it."
+
+"AI AI AI, I think I'm getting a headache."
+
+This article is the first of a series meant to demystify AI, aimed at MDs and other clinicians but without too much medical jargon. We begin with definitions (and indefinitions), with examples, of a few of the most popular terms in the lay and technical presses.
+
+# Artificial Intelligence
+
+There is no generally accepted definition of AI.
+
+This begins a theme that will run throughout this article, best illustrated with an analogy from Humanities 101.
+
+If you pick up a stack of Western humanities textbooks with chronologies of the arts from prehistory to the present, you will likely find a [fairly unified canon](https://en.wikipedia.org/wiki/Western_canon) (try to find a textbook that does _not_ include Michelangelo and Bach, even if it is brand new and socially aware) up until the 1970s or 1980s . At this point, scholarly consensus wanes. It has not had time to mature. As the present day approaches, the selection of important pieces and figures, and even the acknowledgement and naming of new artistic movements (e.g. "[post-postmodernism](https://en.wikipedia.org/wiki/Post-postmodernism)"), becomes idiosyncratic to the specific set of textbook writers.
+
+While the phrase "AI" has been around since [1955](https://aaai.org/ojs/index.php/aimagazine/article/view/1904), the recent explosion in tools, techniques, and applications has destabilized the term. Everyone uses it in a slightly different way, and opinions vary as to what "counts" as AI. This reality requires a certain mental flexibility, and an acknowledgement that any definition of AI (or any of the other terms discussed below) will be incomplete, biased, and likely to change.
+
+With that in mind, we offer three definitions:
+
+
+## General AI
+{:height="400px"}
+
+This is the kind of AI that can reason about any kind of problem, without the requirement for explicit programming. In other words, general AI can think flexibly and creatively, much in the same way humans can. _General AI has not yet been achieved_. [Predictions](https://hackernoon.com/frontier-ai-how-far-are-we-from-artificial-general-intelligence-really-5b13b1ebcd4e) about when it will be achieved range from the next few decades, to the next few centuries, to never. [Perceptions](https://www.newyorker.com/magazine/2018/05/14/how-frightened-should-we-be-of-ai) of what will happen if it is achieved range from salvific to apocalyptic.
+
+
+## Narrow AI
+{:class="img-responsive"}
+
+This is the kind of AI that can perform well if the problem is well-defined, but isn't good for much else. Most AI breakthroughs in recent years are "narrow," algorithms that can meet or exceed human performance on a specific task. Instead of "thinking, general-purpose wonder-boxes," current AI successes are more akin to "[highly specialised toasters](https://aeon.co/ideas/the-ai-revolution-will-be-led-by-toasters-not-droids)." Because most AI is narrow, and quite so, when clinicians see any article or headline claiming that "AI beats doctors," they would be wise to ask questions proposed by radiologist and AI researcher, [Dr. Oakden-Rayner](https://lukeoakdenrayner.wordpress.com/2016/11/27/do-computers-already-outperform-doctors/): "What, _exactly_, did the algorithm do, and is that a thing that doctors actually do (or even want to do)?" A more comprehensive rubric for evaluating narrow AI and planning projects is available in the appendix to Brynjolfsson and Mitchell's [practical guide to AI](http://science.sciencemag.org/content/358/6370/1530.full) from _Science_ magazine.
+
+
+## AI
+
+
+We will finish with Kevin Kelly's [flexible and aware definition](https://ideas.ted.com/why-we-need-to-create-ais-that-think-in-ways-that-we-cant-even-imagine/):
+>In the past, we would have said only a superintelligent AI could beat a human at Jeopardy! or recognize a billion faces. But once our computers did each of those things, we considered that achievement obviously mechanical and hardly worth the label of true intelligence. We label it "machine learning." Every achievement in AI redefines that success as "not AI."
+
+This view of AI takes into account the continual progression of the field, in sync with the progression of humans that produce and use the technology.
+
+Kelly's quote calls to mind George R.R. Martin's [humorously sobering line](http://www.georgerrmartin.com/about-george/on-writing-essays/on-fantasy-by-george-r-r-martin/),
+ >Fantasy flies on the wings of Icarus, reality on Southwest airlines.
+
+If we flip this quote on its head a bit, we can see that real-life flight is a comprehensible thing, intellectually accessible to any person willing to put in time to learn a little physics and engineering, to the point that it becomes banal. Even for those who know nothing of the math and science, most are unmovably bored during their typical commuter flight, some fast asleep even before the roar of the tarmac gives way to the smooth and steady stream at 30,000 feet.
+
+AI is now, in the minds of many, more akin to Icarus than the 5:15 to Atlanta.
+
+In this series we hope to gain the middle ground: help the reader gain and maintain a sense of possibility and perspective, but also understand the mundane ins-and-outs of day-to-day AI.
+
+# Machine Learning
+
+A common definition of ML goes something like,
+>Given enough examples, an algorithm "learns" the relationship between inputs and outputs, that is, how to get from point A to point B, without being told exactly how points A and B are related.
+
+This is reasonable, but incomplete. Each algorithm has its own [flavor](https://xkcd.com/2048/): assumptions, strengths, weaknesses, uses, and adherents.
+
+The simplest example generalizes well to more complex algorithms:
+
+Imagine an AI agent that is shown point A and point B of multiple cases. If it assumes a linear relationship between input (A) and output (B), which is often a reasonable approach, it can then calculate (“learn”) a line that approximates the trend. After that, all you have to do is give the AI an input, even one it hasn't seen before, and it will tell you the most likely output. This describes the basic ML algorithm known as [Linear Regression](https://www.youtube.com/watch?v=zPG4NjIkCjc).
+
+
+
+While linear regression is powerful and should not be underestimated, it depends on the [core assumption](https://xkcd.com/1725/) we outlined, that is, the data are arranged in something approaching a straight line. From linear and logistic regression through high-end algorithms such as gradient-boosting machines ([GBM](https://www.youtube.com/watch?v=OaTO8_KNcuo)s) and Deep Neural Networks (DNNs), each machine learning algorithm has certain assumptions. A major advantage of many newer algorithms is that their assumptions are far more flexible than the classic regression functions available on high school graphing calculators, but at their core they are still abstracted approximations of the real world, equations defined by humans.
+
+Since the goal of this series is to help the reader try out some machine learning with hands-on coding, we should also note here that in most cases, running a GBM is exactly as easy as running linear regression (LR), if not easier: same number of lines of code, same basic syntax. Most of the time you have to change only one or a few words to switch, for example, from GBM to LR and vice versa. Often, an algorithm such as GBM is actually _easier_ to put into play, because it does not place as many requirements on the type and shape of data it will accept ([roughly 80% ](https://www.forbes.com/sites/gilpress/2016/03/23/data-preparation-most-time-consuming-least-enjoyable-data-science-task-survey-says/) of a data science job is collecting and whipping data into something palatable to the algorithm). Complexities may come later, when fine-tuning, and interpreting and implementing findings, but those problems are by no means intractable. We'll get to that in a later post.
+
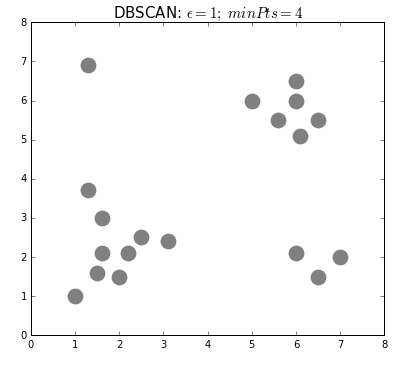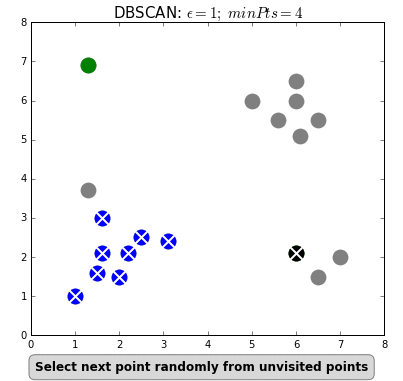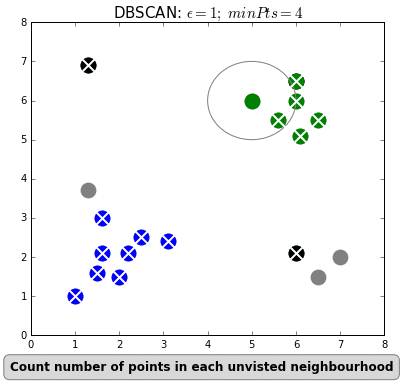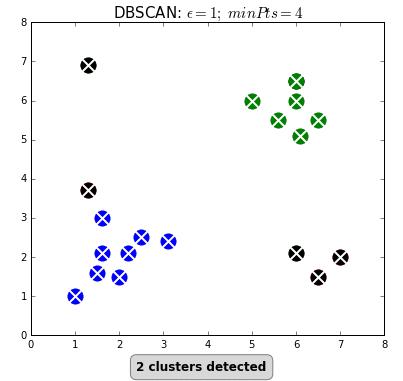
+Even "unsupervised" machine learning, wherein the algorithm seeks to find relationships in data rather than being told exactly what these relationships should be, is based on iterations of simple rules. Below is an example of an unsupervised learning algorithm called [DBSCAN](https://dashee87.github.io/data%20science/general/Clustering-with-Scikit-with-GIFs/). DBSCAN is meant to automatically detect groupings, for example, gene expression signatures or areas of interest in a radiographic image. It randomly selects data points, applies a simple rule to see what other points are "close enough," and repeats this over and over to find groups. You have to choose which numbers to use for `epsilon`: how close points have to be to share a group; and `minPts`: the minimum number of points needed to count as a group. The makers of this GIF chose 1 and 4, respectively.
+
+
+
+As you can see here, "unsupervised" machine learning is clever and useful, but not exactly "unsupervised." You have to choose which algorithm to use in the first place, and almost all algorithms have some parameters you have to set yourself, often without knowing exactly which values will be best (would `minPts = 3` have been better here, to catch that bottom right group?). It is not so different from selecting a drug and its dosage in a complex case: sometimes you will have clinical trials to help you make that decision, sometimes you have to go with what worked well in the past, and sometimes it's pure trial-and-error, aided by your clinical acumen and luck.
+
+Lastly, as always, regardless of the elegance of the algorithm, the machine can only take the data we provide. Junk in still equals [junk out](http://www.tylervigen.com/spurious-correlations), even if it goes through an [ultraintelligent washing machine](https://xkcd.com/1838/).
+
+
+## Deep Learning
+
+Deep Learning (DL) is a subset of machine learning, best known for its use in computer vision and language processing. Most DL techniques use the analogy of the human brain. A "neural network" connects discrete "neurons," individual algorithms that each process a simple bit of information and decide whether it is worth passing to the next neuron. Over time the accumulation of simple decisions yields the ability to process huge amounts of complex data.
+
+
+
+For example, the neural network may be able to tell you [whether or not something is a hotdog](https://medium.com/@timanglade/how-hbos-silicon-valley-built-not-hotdog-with-mobile-tensorflow-keras-react-native-ef03260747f3), what you probably meant [when you asked Alexa to "play Prince,"](https://developer.amazon.com/blogs/alexa/post/4e6db03f-6048-4b62-ba4b-6544da9ac440/the-scalable-neural-architecture-behind-alexa-s-ability-to-arbitrate-skills) or whether the retina shows signs of [diabetic retinopathy](https://doi.org/10.1001/jama.2016.17216). These successes in previously intractable problems led researchers and pundits to claim that DL was the breakthrough that would lead to general AI, but, in line with Kevin Kelly's fluid definition cited above, experience has now tempered these claims with [specific concerns and shortcomings](https://arxiv.org/abs/1801.00631).
+
+
+# Big Data
+
+
+
+The best definition of "big data" borders on the tautological:
+>Data are "big" when they require specialized software to process.
+
+In other words, if you can deal with it easily in Microsoft Excel, your database probably is not big enough to qualify. If you need something fancy like Hadoop or NoSQL, you are probably dealing with big data. Put simply, these applications [excel](https://www.brainscape.com/blog/wp-content/uploads/2012/10/Jj5i1Ge.jpg) at breaking massive datasets into smaller chunks that are analyzed across many machines and/or in a step-wise fashion, with the results stitched together along the way or at the end.
+
+There is no hard-and-fast cutoff, no magic number of rows on a spreadsheet or bytes in a file, and no single "big data algorithm." In general, the size of big data is increasing rapidly, especially with such tools as always-on fitness trackers that include a growing number of sensors and can yield [troves of data](https://ouraring.com/how-oura-works/), per person, per day. The major task is to separate the wheat from the chaff, the signal from the noise, and find novel, actionable trends. The [larger the data](https://www.wired.co.uk/article/craig-venter-human-longevity-genome-diseases-ageing), the more the potential: for finding something meaningful; for drowning in so many meaningless bits and bobs.
+
+# Summary
+
+AI and related terms have no completely satisfying or accepted definitions.
+
+They are relatively new and constantly evolving.
+
+Flexibility is required[^1].
+
+Behind all of the technological terms, there are humans with mathematics and computers, creativity and bias, just as there is a human inside the white coat next to the EKG.
+
+[^1]: A linguistic gem from an early AI researcher is here apropos: "Time flies like an arrow. Fruit flies like a banana." There are a [delightful number of ways to interpret](https://en.wikipedia.org/wiki/Time_flies_like_an_arrow;_fruit_flies_like_a_banana#Analysis_of_the_basic_ambiguities) this sentence, especially if you happen to be a computer. How much flexibility is too much? Too little?
+
+ The paper with the original Oettinger quote was frustratingly hard to find, as is often the case with classic papers from the middle of the 1900s. So save you the hassle, [here's a PDF](http://worrydream.com/refs/Scientific%20American,%20September,%201966.pdf). The article starts on p. 166.
+
+[^2]: Lest you think I've lost perspective on what really matters, here's a [comparison of the Google search trends over time for "Artificial Intelligence" and "potato."](https://trends.google.com/trends/explore?date=all&q=%2Fm%2F0mkz,%2Fm%2F05vtc) Happy Thanksgiving.
diff --git a/2019-06-10-python-write-my-paper.md b/2019-06-10-python-write-my-paper.md
@@ -0,0 +1,132 @@
+---
+layout: post
+title: "Python, Write My Paper"
+toc: false
+image: /images/unsplash-grey-flowerbuds.jpg
+categories:
+ - AI for MDs
+tags:
+ - coding
+ - python
+ - fstring
+ - laziness
+---
+
+{:class="img-responsive"}
+
+Computers are good at doing tedious things.[^1]
+
+Many of the early advances in computing were accomplished to help people do tedious things they didn't want to do, like the million tiny equations that make up a calculus problem.[^2] It has also been said, and repeated, and I agree, that one of the three virtues of a good programmer is laziness.[^3]
+
+One of the most tedious parts of my job is writing paragraphs containing the results of lots of math relating to some biomedical research project. To make this way easier, I use a core Python utility called the `f-string`, in addition to some other tools I may write about at a later date.[^4]
+
+## The problem
+
+First, here's an example of the kinds of sentences that are tedious to type out, error prone, and have to be fixed every time something changes on the back end (--> more tedium, more room for errors).
+
+"In the study period there were 1,485,880 hospitalizations for 708,089 unique patients, 439,696 (62%) of whom had only one hospitalization recorded.
+The median number of hospitalizations per patient was 1 (range 1-176, [1.0 , 2.0])."
+
+The first paragraph of a results section of a typical medical paper is chock-full of this stuff. If we find an error in how we calculated any of this, or find that there was a mistake in the database that needs fixing (and this happens woefully often), all of the numbers need replaced. It's a pain.
+How might we automate the writing of this paragraph?[^5]
+
+## The solution
+
+First, we're going to do the math (which we were doing anyway), and assign each math-y bit a unique name. Then we're going to plug in the results of these calculations to our sentences.
+If you're not familiar with Python or Pandas, don't worry - just walk through the names and glance at the stuff after the equals sign, but don't get hung up on it.
+The basic syntax is:
+
+```python
+some_descriptive_name = some_dataset["some_column_in_that_dataset"].some_mathy_bit()
+```
+
+After we generate the numbers we want, we write the sentence, insert the code, and then use some tricks to get the numbers in the format we want.
+
+In most programming languages, "string" means "text, not code or numbers." So an `f-string` is a `formatted-string`, and allows us to insert code into blocks of normal words using an easy, intuitive syntax.
+
+Here's an example:
+
+```python
+name_of_word_block = f"""Some words with some {code} we want Python to evaluate,
+maybe with some extra formatting thrown in for fun,
+such as commas to make long numbers more readable ({long_number:,}),
+or a number of decimal places to round to
+({number_with_stuff_after_the_decimal_but_we_only_want_two_places:.2f},
+or a conversion from a decimal to a percentage and get rid of everything after the '.'
+{some_number_divided_by/some_other_number*100:.0f}%)."""
+```
+
+First, declare the name of the block of words. Then write an `f`, which will tell Python we want it to insert the results of some code into the following string, which we start and end with single or triple quotes (triple quotes let you break strings into multiple lines).
+Add in the code within curly brackets, `{code}`, add some optional formatting after a colon, `{code:formatting_options}`, and prosper.
+
+As you can see from the last clause, you can do additional math or any operation you want within the `{code}` block. I typically like to do the math outside of the strings to keep them cleaner looking, but for simple stuff it can be nice to just throw the math in the f-string itself.
+
+Here's the actual code I used to make those first two sentences from earlier. First the example again, then the math, then the f-strings.[^7]
+
+"In the study period there were 1,485,880 hospitalizations for 708,089 unique patients, 439,696 (62%) of whom had only one hospitalization recorded.
+The median number of hospitalizations per patient was 1 (range 1-176, [1.0 , 2.0])."
+
+```python
+n_encs = data["encounterid"].nunique()
+n_pts = data["patientid"].nunique()
+
+pts_one_encounter = df[df["encounternum"] == 1].nunique()
+min_enc_per_pt = df["encounternum"].min()
+q1_enc_per_pt = df["encounternum"].quantile(0.25)
+median_enc_per_pt = df["encounternum"].median()
+q3_enc_per_pt = df["encounternum"].quantile(0.75)
+max_enc_per_pt = df["encounternum"].max()
+
+sentence01 = f"In the study period there were {n_encs:,} hospitalizations for {n_pts:,} unique patients, {pts_one_encounter:,} ({pts_one_encounter/n_pts*100:.0f}%) of whom had only one hospitalization recorded. "
+sentence02 = f"The median number of hospitalizations per patient was {median_enc_per_pt:.0f} (range {min_enc_per_pt:.0f}-{max_enc_per_pt:.0f}, [{q1_enc_per_pt} , {q3_enc_per_pt}]). "
+```
+
+If you want to get real ~~lazy~~ ~~fancy~~ lazy, you can combine these sentences into a paragraph, save that paragraph to a text file, and then automatically include this text file in your final document.
+
+```python
+paragraph01 = sentence01 + sentence02
+results_text_file = "results_paragraphs.txt"
+with open(results_text_file, "w") as text_file:
+ print(paragraph01, file=text_file)
+```
+
+To automatically include the text file in your document, you'll have to figure out some tool appropriate to your writing environment. I think there's a way to source text files in Microsoft Word, though I'm less familiar with Word than other document preparation tools such as LaTeX. If you know how to do it in Word, let me know (or I'll look into it and update this post).
+
+Here's how to do it in LaTeX. Just put `\input` and the path to your text file at the appropriate place in your document:[^6]
+
+```latex
+\input{"results_paragraphs_latex.txt"}
+```
+
+With this workflow, I can run the entire analysis, have all the mathy bits translated into paragraphs that include the right numbers, and have those paragraphs inserted into my text in the right spots.
+
+I should note that there are other ways to do this. There are ways of weaving actual Python and R code into LaTeX documents, and RMarkdown is a cool way of using the simple syntax of Markdown with input from R. I like the modular approach outlined here, as it lets me just tag on a bit to the end of the Python code I was writing anyway, and integrate it into the LaTeX I was writing anyway. I plan to use this approach for the foreseeable future, but if you have strong arguments for why I should switch to another method, I would love to hear it, especially if it might better suit my laziness.
+
+Addendum: As I was writing this, I found a similar treatment of the same subject. It's great, with examples in R and Python. [Check it out](https://jabranham.com/blog/2018/05/reporting-statistics-in-latex/).
+
+[^1]: _Automate the Boring Stuff_ by Al Sweigart is a great introduction to programming in general, and is available for free as a [hypertext book](https://automatetheboringstuff.com/). It teaches exactly what its name denotes, in an interactive and easy-to-understand combination of code and explanation.
+
+[^2]: I'm revisiting [Walter Isaacson's _The Innovators"](https://en.wikipedia.org/wiki/The_Innovators_(book)), which I first listened to before I got deeply into programming, and on this go-through I am vibing much harder with the repeated (and repeated) (and again repeated) impetus for building the first and subsequent computing machines: tedious things are tedious.
+
+[^3]: The other two are impatience and hubris. Here is one of the [most lovely websites on the internet](http://threevirtues.com/)
+
+[^4]: For example, TableOne, which makes the (_incredibly_ tedious) task of making that classic first table in any biomedical research paper _so much easier_. Here's a link to [TableOne's project page](https://github.com/tompollard/tableone), which also includes links out to examples and their academic paper on the software.
+
+[^5]: Assign it to a resident, of course.
+
+[^6]: You may have noticed that the name of this file is "results_paragraphs_latex.txt" rather than "results_paragraphs.txt," and that's because LaTeX needs a little special treatment if you're going to use the percentage symbol. LaTeX uses the percentage symbol as a comment sign, meaning that anything after the symbol is ignored and left out of the document. You have to "escape" the percentage symbol with a slash, like this: `\%`. I have this simple bit of code that converts the normal text file into a LaTeX-friendly version:
+
+ ```python
+ # make a LaTeX-friendly version (escape the % symbols with \)
+ # Read in the file
+ with open(results_text_file, "r") as file:
+ filedata = file.read()
+ # Replace the target string
+ filedata = filedata.replace("%", "\%")
+ # Write the file
+ text_file_latex = "results_paragraphs_latex.txt"
+ with open(text_file_latex, "w") as file:
+ file.write(filedata)
+ ```
+
+[^7]: You may have noticed there are two datasets I'm pulling from for this, "data," which includes everything on the basis of _hospitalizations_, and "df," short for "dataframe," which is a subset of "data" that only includes each _patient_ once (rather than a new entry for every hospitalization), along with a few other alterations that allow me to do patient-wise calculations.
diff --git a/2019-10-20-chrome-extensions.md b/2019-10-20-chrome-extensions.md
@@ -0,0 +1,142 @@
+---
+layout: post
+title: "Chrome Extensions I've Known and Loved"
+toc: true
+image: https://source.unsplash.com/xNVPuHanjkM
+categories:
+ - general tech
+tags:
+ - browsers
+ - convenience
+---
+
+A friend recently asked which Chrome extensions I use.
+I've gone through many, and will yet go through many more.
+Here are some I find useful, and some I've found useful in the past.
+
+## Extensions I Have Installed Right Now
+
+### [Vimium](https://chrome.google.com/webstore/detail/vimium/dbepggeogbaibhgnhhndojpepiihcmeb?hl=en)
+
+Navigating a modern web browser is usually done with a mouse or a finger/stylus.
+I like to use the keyboard as much as possible.
+
+Most of my browsing is for a particular piece of information I need for a patient, research project, etc.,
+and I'm going to jump from a text editor to a website and back again very quickly.
+If I can do all of that without leaving the keyboard, I feel much cooler (there may or may not be productivity bonuses).
+
+Vimium uses standard keyboard commands from the text editor Vim for navigating Chrome.
+It works on most pages.
+Vim commands take a little getting used to,
+but once you get the hang of them you'll miss them
+whenever you are deprived.
+
+Some examples:
+- `hjkl` for left, up, down, right (respectively). Seems weird at first, but try it and you'll love it in about 8 minutes. (You can also try some of it on your Gmail now - it has `jk` built in for up and down).
+- `/` for search (one less key press than `ctrl-F`!
+- `f` searches for all links on a page, and gives them a one or two letter shortcut. You type that shortcut, and it activates the link (hold shift while you type the shortcut to open it in a new tab).
+- `x` closes a tab.
+- `gt` Goes to the next Tab. `gT` Goes to the previous tab (think of this as `g-shift-t` and it's intuitive).
+
+### [ColorPick Eyedropper](https://chrome.google.com/webstore/detail/colorpick-eyedropper/ohcpnigalekghcmgcdcenkpelffpdolg?hl=en)
+
+The ability to grab an exact color from a webpage (or PDF) is very useful.
+Maybe you are preparing a presentation and would like to/are forced to use a particular color palette from your institution, or the institution you will be presenting at.
+Maybe you are making figures for an article submission and want to match styling.
+Maybe you would like to color code a document based on an image from an article.
+Whatever it is, it's nice to exactly match a color.
+
+ColorPick Eyedropper is an extension that gives you a crosshair you can place on anything open in Chrome (not just webpages)
+that will give you the code for that color in whichever format you need
+(hex code, RGB, etc.).
+
+### [Loom](https://www.loom.com/)
+
+Loom is free video recording software that can run in Chrome or on a desktop app.
+It's great for simple screencasts, which is all most of us need to get some idea across.
+One of my favorite features is that, if I can run Chrome, I can run Loom - I've recorded little screencasts on locked-down corporate computers.
+
+When I'm thinking about democratized tech,
+I'm thinking about resource-poor constraints
+(e.g. availability of machines, period)
+as well as resource-rich constraints
+(e.g. locked-down machines at your institution,
+that do not let you install or configure much at all).
+Loom is an example of technology that can help in both situations,
+as long as you have a machine modern enough to run Chrome,
+and your admin have at least allowed Chrome on the machine.
+
+
+### [Poll Everywhere for Google Slides](https://www.polleverywhere.com/app/google-slides/chrome)
+
+PollEv is awesome for audience participation in Google Slide presentations.
+You have to have the extension installed for it to work.
+I'm currently giving a lot of presentations, so I have it enabled
+(and will for the foreseeable future).
+
+### Zotero Connector (and Mendeley Importer)
+
+Mendeley used to be my go-to reference manager.
+I moved to Zotero for a variety of reasons
+(mostly: it's free, open-source, and therefore quite extensible.
+Mendeley, for example, could not approve a Sci-Hub integration.
+If you don't know what Sci-Hub is, Google "zotero sci-hub" and let me know if you are inspired or enraged, or both.).
+
+The Zotero extension both allows you to save references to your Zotero library,
+and to use Zotero plugins for reference management in Google Docs.
+
+### Chrome Remote Desktop
+
+Remote desktops are cool. Chrome's built-in solution is very serviceable.
+
+## Extensions I've Known and Loved
+
+### KeyRocket for Gmail
+
+KeyRocket is a suite of tools that help you learn keyboard shortcuts to navigate a variety of software.
+This is a Gmail extension that does just that.
+Whenever you do a thing manually (i.e. by clicking around),
+KeyRocket will put up a brief, inobtrusive popup
+(that you don't have to click on to exit)
+showing you how you could have done that with a keyboard shortcut.
+
+When you stop getting so many popups, you have become proficient with the shortcuts. At some point, you deactivate the extension and bid it a fond adieu.
+
+### [Librarian for arXiv](https://blogs.cornell.edu/arxiv/2017/09/28/arxiv-developer-spotlight-librarian-from-fermats-library/)
+
+arXiv.org is a great resource for finding and disseminating
+research, particularly in computational sciences and related areas.
+Librarian is an extension that makes getting the citations and references very easy.
+
+### [Unsplash Instant](https://chrome.google.com/webstore/detail/unsplash-instant/pejkokffkapolfffcgbmdmhdelanoaih?hl=en)
+
+This replaces your new tab screen with a randomly chosen image from Unsplash.
+Unsplash.com is a wonderful resource for freely usable, gorgeous images.
+I give a lot of presentations, so I use Unsplash for finding pretty backgrounds and illustrative images.
+It is nice to have a pretty new tab screen, and it's a nice way to find images to archive for future use (you can just click the heart icon, or manually download).
+
+P.S. Most of the splash images on my website are from Unsplash (e.g. the one at the left of this post if you're on a large screen, or at the top if you're on a small screen).
+
+I might just reactivate this extension. I'm not sure why I deactivated it.
+
+### [Markdown Viewer](https://chrome.google.com/webstore/detail/markdown-viewer/ckkdlimhmcjmikdlpkmbgfkaikojcbjk?hl=en)
+
+I think it is clear that I love Markdown.
+Markdown Viewer is an extension that allows you to preview
+what Markdown will look like when it's rendered,
+either from a local file or from a website.
+
+Now that I'm writing more Markdown, I might also turn this one back on.
+
+P.S. I turned it back on. It's awesome.
+
+### Others...
+
+These are just the ones I had disabled, but not uninstalled.
+I know I've used others, and I'm sure many of them were cool.
+My laziness at the moment is such that I'm not going to go digging around.
+
+## What about you?
+
+Do you have extensions you know and love, or have known and loved?
+Let me know, and I'll check them out.
diff --git a/2019-10-20-r-markdown-python-friends.md b/2019-10-20-r-markdown-python-friends.md
@@ -0,0 +1,86 @@
+---
+layout: post
+title: "R Markdown, Python, and friends: write my paper"
+toc: true
+image: https://source.unsplash.com/OfMq2hIbWMQ
+categories:
+ - AI for MDs
+tags:
+ - coding
+ - software
+ - laziness
+ - "R Markdown"
+ - Python
+ - Markdown
+ - "academic writing"
+---
+
+## R Markdown is my spirit animal
+
+In a [previous post]({% post_url 2019-06-10-python-write-my-paper %}) I talked about how easy it is, if you're already doing your own stats anyway in some research project, to have a Python script output paragraphs with all the stats written out and updated for you to add into your paper.
+
+The main problem with the approach I outlined was how to get those nicely updated paragraphs into the document you are sharing with colleagues.
+
+Medicine, in particular, seems wed to Microsoft Word documents for manuscripts. Word does not have a great way to include text from arbitrary files, forcing the physician-scientist to manually copy and paste those beautifully automated paragraphs. As I struggled with this, I thought (here cue Raymond Hettinger), "There must be a better way."
+
+Turns out that better way exists, and it is R Markdown.
+
+Though I was at first resistant to learning about R Markdown, mostly because I am proficient in Python and thought the opportunity cost for learning R at this point would be too high, as soon as I saw it demoed I changed my tune. Here's why.
+
+## Writing text
+- R Markdown is mostly markdown.
+ - Markdown is by far the easiest way to write plaintext documents, especially if you want to apply formatting later on without worrying about the specifics while you're writing (e.g. `#` just specifies a header - you can decide how you want the headers to look later, and that styling will automatically be applied).
+ - Plaintext is beautiful. It costs nearly nothing in terms of raw storage, and is easy to keep within a version control system. Markdown plaintext is human-readable whether or not the styling has been applied. Your ideas will never be hidden in a proprietary format that requires special software to read.
+ - I had been transitioning to writing in Markdown anyway, so +1 for R Markdown.
+- R Markdown is also a little LaTeX.
+ - LaTeX is [gorgeous](https://tex.stackexchange.com/questions/1319/showcase-of-beautiful-typography-done-in-tex-friends) and wonderful, the most flexible and expressive of all the typesetting tools (though not as fast as our old friend Groff...). It also has a steeper learning curve than Markdown, and is not so pretty on the screen in its raw form. R Markdown lets you do the bulk of your work in simple Markdown, then seamlessly invoke LaTeX when you need something a little fancier.
+- R Markdown is also a little HTML.
+ - HTML is also expressive, and can be gorgeous and wonderful. It is a pain to write. As with LaTeX, you can simply drop in some HTML where you need it, and R Markdown will deal with it as necessary.
+- R Markdown is academic-friendly.
+ - Citations and formatting guidelines for different journals are the tedious banes of any academic's existence. R Markdown has robust support for adding in citations that will be properly formatted in any desired style, just by changing a tag at the top of the document. Got a rejection from Journal 1 and want to submit to Journal 2, which has a completely different set of citation styles and manuscript formatting? NBD.
+
+## Writing code
+R Markdown, as the name implies, can also run R code.
+Any analysis you can dream of in R can be included in your document, and you can choose whether you want to show the code and its output, the output alone, or the code alone.
+People will think you went through all the work of making that figure, editing it in PowerPoint, screenshotting it to a .png, then dropping that .png file into your manuscript, but the truth is...
+you scripted all of that, so the manuscript itself made the .png and included it where it needed to go.
+
+R Markdown is by no means restricted to R code.
+This is the killer app that won me over.
+Simply by specifying that a given code block is Python,
+and installing a little tool (`reticulate`) that allows R to interface with Python,
+I can run arbitrary Python code within the document and capture the output however I want.
+That results paragraph? Sure.
+Fancy images of predictions from my machine learning model? But of course.
+
+If you don't want to use any R code ever, that's fine. R Markdown doesn't mind.
+Use SAS, MATLAB (via Octave), heck, even bash scripts - the range of language support is fantastic.
+
+## Working with friends
+R Markdown can be compiled to pretty much any format you can dream of.
+My current setup simultaneously puts out an HTML document (that can be opened in any web browser), a PDF (because I love PDFs), and (AND!) a .docx Word file,
+all beautifully formatted, on demand, whenever I hit my keyboard shortcut. I can preview the PDF or HTML as I write, have a .docx to send to my PI, and life is good.
+
+Also, because you can write in any programming language, you can easily collaborate between researchers that are comfortable in different paradigms.
+You can pass data back and forth between your chosen languages (for me, R and Python),
+either directly or by saving intermediate data to a format that both languages can read.
+
+## Automating tasks
+Many analyses and their manuscripts, especially if they use similar techniques (e.g. survival modeling), are rather formulaic.
+Many researchers have scripts they keep around and tweak for new analyses revolving around the same basic subject matter or approach.
+With R Markdown, your entire manuscript becomes a runnable program, further automating the boring parts of getting research out into the open.
+
+One of the [first introductions](https://www.youtube.com/watch?v=MIlzQpXlJNk) I had to R Markdown shared the remarkable idea of setting the file to run on a regular basis,
+generating a report based on any updated data,
+and then sending this report to all the interested parties automatically.
+While much academic work could not be so fully automated, parts of it certainly can be.
+
+Perhaps your team is building a database for outcomes in a given disease, and has specified the analysis in great detail beforehand.
+One of my mentors gives the advice that in any project proposal you should go as far as to mock up the results section,
+including all figures,
+so you make sure you are collecting the right data.
+If this was done in an R Markdown document rather than a simple Word document,
+you could have large parts of the template manuscript
+become the real manuscript as the database fleshes out over time.
+Then when it's done, look over the data, make additions and subtractions as needed,
+write the discussion sections, and send it in.
diff --git a/2020-03-13-nih-get-you-funded.md b/2020-03-13-nih-get-you-funded.md
@@ -0,0 +1,314 @@
+---
+layout: post
+title: "Notes on the book 'How the NIH Can Help You Get Funded'"
+toc: true
+image: https://source.unsplash.com/OfMq2hIbWMQ
+tags:
+ - "physician-scientist"
+ - "academic writing"
+ - funding
+ - "career development"
+---
+
+# How the NIH Can Help You Get Funded
+
+[Mike Becich](https://www.dbmi.pitt.edu/person/michael-j-becich-md-phd) is a wonderful researcher and entrepreneur, and chairman of the Department of Biomedical Informatics at the University of Pittsburgh. I met him on the residency and fellowship interview trail, and, among other things, we talked about grant strategy and how to build a career doing research in biomedical machine learning/informatics/data science/etc. He was kind enough to give me a copy of the book <ins>How the NIH Can Help You Get Funded</ins>, which is exactly what it sounds like: a how-to and strategy book for academic research scientists, most of whom depend on funding from the National Institutes of Health to support their work and salaries. The book is a publication of [Oxford University Press](https://global.oup.com/academic/product/how-the-nih-can-help-you-get-funded-9780199989645?cc=us&lang=en&) and also available from [Amazon](https://www.amazon.com/How-NIH-Can-Help-Funded/dp/0199989648).
+
+These are my notes from and synthesis of this book. The synthesis was made by going through my notes and extracting things I thought were key and may be useful for quick review; the full notes I took as I was reading are copied below that.
+
+If you notice any mistakes, misconceptions, or flatout wrongheadedness, please shoot me an email. Also, though the general outline of how the NIH works is fairly stable, specifics change from year to year and this book, as with almost every book of its genre, was outdated upon publication. It will be at least a few years before I apply for funding, at which point an even larger number of specific details will be out of date, so what I am looking for is general strategy and key history.
+
+# Synthesis
+This book is a bit hard to summarize, as it has a mix of very practical and technical information, with general guidance, strategy, and advice (to use the parlance du jour, strategy and tactics). I figured this would be the case going in, hence this public store of notes that I can quickly review.
+
+Some general things:
+- The NIH is about science, but is run by, and full of, people. These people have personalities, hopes, fears, egos, soft spots, families, and digestive systems. If you keep this front of mind as you communicate your science, you'll have a Good Time.
+- Of all the people at the NIH, your main points of contact are the Program Officers (POs). It is a completely sane and compassionate approach to try and make their lives easier. Also realize that their job is to build a portfolio of cool and important work for their organization, and that they often had prior lives as PIs themselves. So do your homework, know what stresses and timelines they are under, and do your best with your science and your personal communication to develop a relationship of mutual respect and care.
+- Also think of your proposal itself as building a relationship with all its readers - you want to be nice to them (make it easy to read and rate - many specific tips and tricks herein), and to make it exciting and fun to imagine what success will look like for your project, the team, the institution that is considering sponsoring it, the patients who will benefit, and the world at large. (Don't resort to corporate technobabble BS, as this makes it less fun/very irritating to read).
+- The NIH is also a government organization, and is therefore subject to all manner of red tape, budget constraints (and cuts), fickle bureaucrats, and delays. If you know the timelines and possibilities going in, you'll have a Better Time than You Would Otherwise.
+- The R01 Holy Grail makes more sense to me now, since going through this book while also talking to friends who are early in their careers as physcian-scientists. The grant is attached to you, not your institution. You have a large amount of freedom to do what you want with it (with the exception of a small number of funding mechanisms, the NIH pretty much gives you money and gets out of your way - you have to show productivity, but exactly how you accomplish that is your choice). It can be renewed, and this renewal is a bit easier if you're an early investigator. If you want to be a physician-scientist, having funding will let you lighten up on the patient load. You can also hire brilliant people, and having the R01 is a draw for top talent.
+- R01 is a pretty good way to fill 2/3 of the triumvirate of what makes careers happy: competence/expertise (you are by definition an expert in your field), autonomy, and relatedness (you bring this - a sense of connection to the people you work with and for, and the cause).
+- The "publish or perish" thing makes more sense to me now as well. Tenure seems to be going away at many institutions, or is veryveryvery difficult to get, so a researcher's life is a never-ending cycle of applying for grants that have a government-mandated average length of 4 years. If you want a renewal, or a successful new grant, you have to show productivity. Though there is some understanding that certain kinds of work take a long time to bear fruit, you basically have to have first and/or last author pubs in decent-to-good journals rolling out with some consistency to remain competitive. This book helped me own my identity as a physician-scientist (in training), and all the trappings that go with it, including the high likelihood that I will live most of my career on these funding cycles.
+- Speaking of owning my career, the book also makes me curious about non-NIH funding mechanisms, e.g. through other govt orgs or third parties. This book mentions some, including the Dept of Defense and National Science Foundation. If anyone has suggestions for places I should check out that may be conducive to a research career in bioinformatics/data science/machine learning for medicine, particularly blood cancers, with major interests in healthcare disparities and community/population health, shoot me an email. I also want to see patients, so certain industry careers may be out - but I know that industry is also a complex and varied space, with as many shapes of careers as there are individuals, so would be open to hearing about those avenues as well.
+- As I finished this book I was also overwhelmed with gratitude to my undergraduate alma mater, Brigham Young University, for giving me singular opportunities for research across the institution that led me to a career in research, to my medical school alma mater, Cleveland Clinic Lerner College of Medicine, for setting me up to be successful, by giving me time and headspace to think about which skills I wanted to develop, plenty of support to get a head start on them, and mentors in every conceivable area. Next, I am so, so excited and grateful for Vanderbilt University Medical Center and the Harrison Society for welcoming me into their fold. The clinical training will be fantastic, and the way they set up their physician-scientists for success (== R01 and other funding mechanisms) occupies a truly rarefied place among academic institutions in the United States. I'll admit I'm scared of the publish-or-perish thing still, but knowing there is a staggering level of support and deep precedence for success helps assuage the imposter syndrome and insecurity.
+- Overall, I'm glad I read this book, and am grateful to Becich and other mentors and friends in science and medicine for getting me to think about the arc of my career and how to increase the likelihood of success. The timing was great (Spring 2020), as I will be entirely engaged in the practice of clinical medicine for the next 2-4 years before jumping back into research. I will likely revisit this book or its successor, as well as these notes, as I scan the horizon for opportunity and learn, as I practice clinical medicine, where help is needed most.
+
+# Notes
+
+## National Institutes of Health
+This chapter provides a background on the NIH.
+- NIH = 27 Institutes and Centers (ICs)
+- began in 1937 with the National Cancer Institute (NCI), then in 1938 cornerstone laid for NIH campus
+- part of executive branch, but Congress authorizes and appropriates funding
+- budget grew ~2x from 1998-2003, but then failed to keep pace with inflation or dropped. This is problematic because ICs would like to plan for multiple years ahead and provide stable commitments to researchers doing important but longer-than-a-year research. The payline (the score and percentile at which grants are funded) is subject to variations in appropriations.
+- budget allocation (rough percents): 80% extramural funding, 11% intramural funding, 5% salaries/admin, 2.5% formal training programs (80+11+5+2.5 = 98.5%)
+- Program Officers (POs) are extramural staff associated with each I/C. The authors make the point first here, and repeatedly throughout the text, that *applicants should contact their PO at each stage of the application, review, and award process*.
+- Office of the Director: coolest thing here is the Office of Strategic Coordination's Common Fund Programs, including the NIH Director awards. These fund cross-cutting, high-risk, possibly high-yield projects (see [here](https://commonfund.nih.gov/highlights) for current highlights), touting the program as the "venture capital" space within the NIH. Some of the data science-y stuff funded by the NIH was/is via the Common Fund, e.g. development of a [data commons](https://grants.nih.gov/grants/guide/notice-files/NOT-RM-17-031.html). The language is purposefully obtuse, matching the tone of Silicon Valley VC, as I think it has to be when they are looking for (again, purposefully) underspecified opportunity to fund interesting work that doesn't quite fall under any IC purview but has potential to be generally useful.
+- A "grant" is different than a loan, contract, or cooperative agreement. The idea is for the IC to determine if your proposed project fits within their strategic plan to have "significant and lasting impact on the field and public health," and then they get out of the way to let you carry out your plan as you see fit, and evaluate your productivity and promise as you apply for renewals, etc.
+ - This jives with what I learned early on in anthropological/ethnomusicological research: it is widely understood that everything might (will) change when you get in the field, but funding and approval bodies need to see evidence of innovation, possible impact, and careful thought before they hand out money and rubber stamps. You then have freedom to do your work as best you know how, and react to unexpected realities as you see fit.
+ - Contracts are for specific needs (e.g. developing an animal model, operating a facility); cooperative agreements are similar to grants but involve substantial "scientific or programmatic" involvement from the federal government (i.e. they are less independent).
+- At this point, the rest of the book is summarized and a timeline is given. The timeline is a concordance of the timelines of Congress, the ICs, the principle investigator (PI), and the standard funding opportunity announcements.
+
+## Institutes and Centers
+- More on the program officer (PO).
+ - POs often were investigators in the field, and have a deep interest in its success.
+ - They see the PIs as "their investigators"
+ - It's ok and encouraged to contact POs in the several ICs that might cover your work, and work with them to determine strategy at every key point along the way
+ - ICs are differents, and POs are different, so don't expect your friend's experience to necessarily match your own
+- Advisory Councils
+ - main job is to determine how well the proposed research fits with the mission of the IC, and if the scientific review was adequate. They don't say much about the scientific merit of a project, as that is the job of the review.
+ - also review concepts for future initiatives, and *cleared concepts* are the fodder for Requests for Applications - keep the pulse on cleared concepts, and you can get a head start on preparing your applications
+- ICs
+ - a list of each IC and key data is presented. Some notes on ideas I found interesting:
+ - Love this sentence on clinical vs. basic science research "The NHLBI believes all the clinical research should have some return on investment, unlike basic science research, which should be designed to explore, not build, and in which return decades later is not uncommon." For one recent example, the Shapley value was developed in the 1950s-60s, but was too computationally expensive for all but rather restricted use, and now undergirds some of the most important paradigms in explainable machine learning systems (SHAP was published in 2017, IIRC). I think we have an uneasy relationship with this concept, that basic and theoretical researchers should essentially be encouraged to play, and play their best, knowing full well that much of the research will never be particularly useful, that some of what will be useful will not be useful for decades, and that it is very, very difficult to determine beforehand which is which. The embrace of intellectual freedom and creativity always butts up against funding constraints, and people want ROI.
+ - NCI R01 success rates financial year 2012: 12.5% for new, 29.4% for renewals, 11.8% for supplement.
+ - NHLBI R01 success rates in financial year 2012: 13% for new, 25.4% for renewal, 33.3% for supplement.
+
+## Center for Scientific Review and the Peer Review Process
+- "Scientific review groups" is the official name for the colloquially known "study sections"
+- Some alphabet soup decoded:
+ - P - program project/center
+ - U - cooperative agreement
+ - T - training
+ - K - career development
+ - N - contracts
+ - F - fellowship
+- How to get your application rejected quickly:
+ - Don't have a cover letter from the IC approving the submission if your budget exceeds $500,000 direct costs in any 1 year
+ - Don't comply with the formatting restrictions
+ - Have the wrong budget type (modular vs. detailed, depending if >/< $250k)
+ - Ignore specific requirements for your application type
+ - Fail to pass the sniff test for A2 applications (revision of an unfunded A1) - apparently they use a combination of NLP and a manual review process to find out if you're just swinging away without changing bats
+- A0/A1/A2
+ - A0 - first/new application
+ - A1 - first revised/amended application
+ - A2 - second revised application, no longer allowed (max attempts = A1)
+ - To convert A2 to A0:
+ - 2/3 of aims must be new
+ - research plan must be substantively changed
+ - OR submit under new mechanism OR in response to an RFA (request for applications) if it happens to match your science
+- Application ID number decoded:
+ - Ex: 1R01 CA1234567-01 A1; 5R01 GM000091-62
+ - First example: New (type I) R01, in the NCI (CA), number 1234567, first year of funding (01), first amendment (A1)
+ - Type 1: new; Type 2: competing renewal; Type 3: competitive supplement; Type 5: noncompetitive supplement
+ - Second example: Noncompeting renewal (type 5) RO1, in the National Institute of General Medical Sciences (NIGMS, abbrev GM here), number 91, in the 62nd year of funding. Cool example - was for the "Structure and Function of Enzymes---Role of Metals." Zinc fingers ftw?!
+- Study section assignment tip: almost always go with the one they give you, not necessarily the one you wanted. Their interest is in getting you a good review, and know the ins and outs likely better than the individual PI.
+- Check the [CSR](https://public.csr.nih.gov/StudySections) website and [RePORTER](https://projectreporter.nih.gov/) to find out which study groups have which interests.
+ - The CSR website gives more granular details on the study section topics, RePORTER gives results and other data, so CSR -> RePORTER is the usual workflow (though I could imagine reasons for RePORTER -> CSR)
+- Also ask your program officer, mentors, and colleagues which study sections you should target.
+- **Think about your study sections at the outset**
+ - If your audience is specific, you can save space and write directly to their knowledge and interests.
+ - This extends all the way to: know their names and what they've published, and create your steel-man arguments (and a little [Carnegie-esque](https://creativesamba.substack.com/p/feeding-the-baby-rabbits) baby rabbit/Edgar Thomson Steel Works action) by citing their work.
+ - Also make sure your work gets in front of them and stuck in their heads with some "presuasion" techniques: present at the conferences they are at, invite them to your seminar series, get their take on what is important in their field now and going forward. Just don't bring up any specific proposals (i.e. don't break the rules, but feel free to water the field).
+- **Hours spent in different parts of the review process** (this is huge):
+ - You spent: 100s of hours
+ - Primary reviewer spends: several hours
+ - Secondary reviewer: less than primary reviewer
+ - Unassigned panel members: zero hours to a few minutes (maybe read abstract and specific aims)
+ - Panel discussion: 10-15 minutes
+ - SO be kind, it's an elevator pitch not a thesis. ?Read [Brief](https://thebrieflab.com/book_brief/) again?
+
+## Getting at Mechanism
+Aha! This is part of why I picked this book. I've always been curious about the specific mxns the NIH uses, and when to use which.
+- R01
+ - ~1/2 of extramural NIH funding is R01
+ - up to 5y length - average length mandated to be 4y, and new and early stage applicants more likely to get 5y award
+ - when you are no longer early or new, then proposing work with shorter time frames is a bit sexier and more likely to get funded
+ - renewable
+ - favors early state investigators (reviewed in their own group, also can reapply within the same funding cycle)
+- R21
+ - "starter" grant
+ - success rate actually lower than R01, d/t large number of increased applications
+ - NIH recommends that the R21 *not* be used as an entry for junior investigators, because:
+ - cannot be renewed
+ - generally limited to ~$275k over 2y
+ - no payline break for new or early applicants
+ - kinda seems like one to avoid, overall - what are the upsides? Not clear from this reading.
+- R03
+ - "Small research grant"
+ - $50k per year for 2y, nonrenewable
+ - time limited, focused - e.g. collect pilot data, perform data analysis, develop assay or model
+ - sometimes limited to K awardees and new investigators
+ - sometimes used to respond to reviewer concerns for R21s, so it would be a (long path) R21 application -> R03 application -> R03 award -> R01 application and award
+- R13
+ - Conference awards, supports specific costs, usually less than $20k (???)
+ - Needs letter from IC R13 PO
+ - 60% success rate
+- R15
+ - Academic Research Enhancement Award (AREA)
+ - Limited to academic components w/in institutions with < $6m in funding from >=4 of the past 7y.
+ - $300k direct costs over 3y, renewable
+ - Seems like a mini-R01 for early researchers at underfunded/newer programs
+- R33
+ - 4 offered per year.
+ - Phase II for successful R21.
+ - Often solicited by funding opportunity announcements and ICs.
+ - Milestone driven (must be quantifiable) w/ Gantt chart or similar timeline
+- R34
+ - Clinical Trial Planning Grant (phase III trials)
+ - E.g. data collection tools, manuals, recruitment strategies, data sharing and multiple IRB submissions if multisite, also pilot studies
+
+Small business grants
+- Small Business Innovation Research (SBIR) R41 and R42
+ - >=40% of the work performed by small business, >=30% by a nonprofit research institution, rest either split or include a third party
+- Small business Technology Transfer (STTR), R43
+ - Primary PI primarily employed by small business. >=67% of the work performed by small business if phase I, >=50% if phase II.
+
+P grants
+- Program projects (P01) or Center (P20, P30, P50, P60)
+- Themes that would benefit from collaboration, and would be unlikely to be successful otherwise
+- Emphasis on supporting new and nontraditional researchers
+- Could be basic or have a clinical component
+- Need a large number of R01 and other awards to justify, and if you are interested in starting one of these work closely with the PO
+
+K awards
+- Career Development Awards
+- Setup:
+ - Award is both for the work itself (e.g. specific aims) and to prepare the investigator for R01, so write it this way - emphasize how the training and mentorship will not only make the project more likely to succeed, but will also set you up for a successful career in your field
+ - This sounds a lot like the ASH awards, with similar strategies:
+ - make sure your primary advisor is going to be an actual advisor, not just a big name on the proposal (the reviewers will probably know if this is the case, and you would be seemingly paradoxically less likely to get the award)
+ - go ahead and have that big name as a secondary advisor/part of an advisory panel, and use their expertise to get a broad view of the field and how to succeed in it (if they have time for specific mentoring, gravy)
+ - write in the first person, about *your* career goals and accomplishments thus far
+ - start the Candidate section with your long-term goals, to frame the conversation and show the reviewers that you are going somewhere
+ - acknowledge your gaps and show how the K award will help you fill them - if you have no major gaps, go for the R01, not the K
+ - have great letters from your mentor and advisory team, that you participate in writing, showing a personalized recommendation and plan, and the qualifications of the mentor(s). Make sure the details in the letters all jive.
+ - Have some first author papers, to the extent possible (reviewers are understanding if you are a clinical scientist and stuff takes forever to get to the manuscript phase)
+ - A collection of poster presentations that never -> manuscript is a bad look
+ - Review papers are +/- helpful, depends on the journal, but might be useful to show commitment to and understanding of the field
+- The grant itself:
+ - Less is more, be ambitious and innovative but realistic about what can be accomplished in the time frame
+- All the Ks:
+ - Different ICs use the same codes for different things, so check the K Kiosk at the NIH website.
+ - But generally:
+ - K08, K23, K24: applicants with clinical degrees
+ - K01, K02: basic scientists
+ - K01, K08, K23, K25: junior investigators
+ - K02, K05, K18, K24: established investigators
+ - K22, K99/R00 (kangaroo!): postdocs (also needs academic appt during award, and need to submit R01 before end of 2nd year of the 3y award). K99 while a postdoc, then R00 activated when faculty appt received
+ - K12: clinical research career dev, established PI submits application to NIH then trainees compete for slots. Coupled with intensive training and resources (similar to the PSTP programs, with biostats support, grant-writing committees and extensive pre-review, etc.)
+ - KL2: similar to K12, Clinical and Translational Science Award (CSTA)
+
+F and T awards
+- F: fellowship, T: training
+- Like Ks, but required to pay back 1y of support through research and teaching (not patient care).
+- Can be obtained predoctoral (e.g. MD-PhDs in training - up to 6y) or postdoctoral - up to 3y.
+
+Summary: Tons of alphabet soup. Most of us will focus on R01 and ways to get to R01 (K,F,T, maybe R21/R03). For more specific stuff, it's always changing anyway, so look it up and talk to your PO.
+
+## Telling Your Story Well
+- Specific Aims may be the only page everyone reads, and then likely cursorily, so grab attention and cut the cruft
+- One way to cut the cruft: know your audience, give only the background that is absolutely necessary and unlikely to be known/agreed upon by all
+- The reviewer is going to be writing bullet points for the summary statement - make this easy for them, do all the work so they don't have to
+- How important, relatively, are the sections of the report?
+- Table 8.1 gives the correlation coefficients bw overall impact score and 5 criterion scores for 2010. The ones that are likely to be of most interest to me:
+
+| Institute | Approach | Significance | Innovation | Investigator | Environment | N |
+| ----------- | ---------- | -------------- | ------------ | -------------- | ------------- | ------ |
+| NCI | 0.80 | 0.67 | 0.59 | 0.53 | 0.45 | 5396 |
+| NHLBI | 0.82 | 0.67 | 0.64 | 0.56 | 0.48 | 3157 |
+
+- So Approach is most correlative, then Significance. In other words, it appears that the way you are doing something is the most important thing (though note that every part of the score is indeed part of the score).
+- On specific aims:
+ - write it first, and rewrite it often
+ - Should address:
+ - why the work is important
+ - why are you the one to solve it
+ - what problems you will solve
+ - how you will do so
+ - what the impact will be
+ - Send a bare-bones version to the PO and colleagues, and take note of the questions they ask
+ - Narrative order (pretty standard): 1. significance of problem, +/- prior contributions of author if available, 2. your take on the problem, lead the reviewer to your hypothesis and approach by integrating prior work and prelim data, 3. hypothesis and overall approach
+ - Picture/diagram/table: can be very useful, but don't just repeat the text, and don't make it harder to understand than just reading the proposal.
+ - One problem with overambitious aims: the reviewers will know that you probably have no idea how long it takes to actually [establish an animal model, recruit patients, build the software, etc.] and you are seen as not having sufficient experience
+ - Aims complementary but not conditional
+ - Generate useful data whether or not your hypothesis is confirmed
+ - **check out the sample R01s from the NIAID** - the link in the book is old, but here's a PDF of the Wahlby grant applicaiton they mention (Image analysis for C elegans) - [Wahlby](https://emergencymed.arizona.edu/sites/emergencymed.arizona.edu/files/sample_nih_proposal_4.pdf), cool for me bc it's fairly ML/AI, and here's the Ratner application, notable for using a schema to communicate the experimental flow - [Ratner](https://www.niaid.nih.gov/sites/default/files/ratnerfull.pdf). Because these were kind of hard to find, I put copies at the following links:
+ - [Ratner PDF](/assets/pdf/ratnerNIHproposal.pdf)
+ - [Wahlby PDF](/assets/pdf/wahlbyNIHproposal.pdf)
+ - 
+ - Can also conclude with a paragraph "Overall Impact" summarizing what you'd like the reviewer to use as the take-home message in their presentation. "..what will be possible after your research has been completed that is not possible or known now."
+
+- On approach:
+ - modular approach based on aims
+ - briefly restate rationale and hypothesis, integrate relevant prelim data, summarize design and individual experiments, conclude with analysis, interpretation of results, potential problems, alternative approaches.
+ - Consider an illustration
+ - key methodology, mentioned by name as needed, but avoid minutiae (it seems that knowing what is and isn't significant is another sign of experience and thoughtfulness).
+ - cite pubs, especially if you wrote them, demonstrating feasability
+ - have good stats - consult with your statistician early and often, and have them review before the study section sees your proposal
+ - indicate how you will interpret the data, especially if they are surprising, and what you will do if the data ends up more fuzzy than expected. I wonder if it would be useful to do what Sekeres recommends and mock up the tables and figures beforehand, to make sure you're collecting the right stuff and analyzing it the right way? (especially if you use the Rmd approach that builds on Sekeres', of actually writing the code that makes the figs, and populating them with mock data, to be filled in as the real stuff comes in)
+ - Consider the proposed work as part of a 10y plan - for you, and for the reviewers (given that renewing R01 is a thing you will pursue and they will encourage). It's not a single project, but a piece of a career.
+
+- On significance:
+ - Significance assumes your work was successful - will the field substantively change as a result? Why does your project deserve funding?
+ - consider splitting into sections: Importance of the Problem, Knowledge to Be Gained/Impact on the Field
+ - know your reviewers, and freely cite their papers as appropriate - they probably think their own work is important and impactful, and you want them nodding along in agreement, if not pounding the table with "Amen!"
+
+- On innovation:
+ - why is your work different and better than the current approaches?
+ - methods do not need to be innovative, if their application is likely to yield innovative findings
+ - consider splitting into sections: Technical Innovation, Conceptual Innovation
+ - another good place for a bullet point, feed the reviewer their line at the meeting
+
+- On the introduction:
+ - If it's an A1, first page will be response/rebuttal.
+ - Remember the reviewer is always right - tell your friends your visceral reaction, write it down, then shred it, rinse and repeat a few times until you've cooled off.
+ - Remember the reviewers that see your rebuttal may be different than the initial reviewers
+ - Take on the big issues, leave the minor stuff - focus on the Resume and Summary of Discussion (individual critiques may have been resolved in the discussion and not revised in the written doc)
+ - "Sometimes your harshest critics can become your biggest fans on resubmission" - and how. I've experienced this repeatedly.
+ - Quote verbatim
+ - Use bullets and indentations to delineate comments and responses
+ - *brief* appreciative acknowledgement, then get to the meat (obsequiousness is ugly)
+ - make sure you are addressing the actual concerns, run it by your PO and friends
+
+- Other sections
+ - Protection of Research Subjects is another area in which your goal is to comfort the reviewers, assure them you know what you're doing (i.e. a great PoRS is unlikely to get you a top score, but a bad one is a red flag).
+ - included in this is your sample size calculation - if not done, or done poorly, will reflect poorly. Biostatisticians are your friends, and will be on the review committees.
+ - Make each biosketch support the overall narrative. Don't copypasta. Seems that the PI (me) should be willing to write/rewrite others' biosketches for jiviness, and send for approval (a la letters of support)
+ - Budgets either modular (module = $25k) or detailed. Detailed if project >$250k, involves foreign institutions, or is one of a few specific mxns that always require it
+ - a too-low budget is also often a sign that you don't know what you're doing
+
+- On writing in general
+ - Use language from the criteria in your section subheadings to make it easy for your reviewers - imagine they have a list of checkboxes they are going through. It's much better to not be creative here, so they can find what they are looking for
+ - Think pagewise: what 2-3 points should no one miss on this page? Consider (sparing) use of bold, italic, etc., but favor white space and emphasis by subtraction.
+ - q: which fonts *does* the NIH allow?
+ - page 123: nice list of how to structure writing for max impact (i.e. use psychological principles: make the first word the subject if possible, put the impactful thing at the end of the sentence, build schema through introductory and summary sentences, etc.)
+ - writing in first person is often less wordy and creates more excitement, buy-in, so use it! (I know there's some difference of opinion on this one, but I'm into bringing personality back to academic and scientific writing, so count me in)
+ - omit needless words. Use modifiers if they have meaning (e.g. "weakly fluorescent").
+ - Don't say your work is "innovative." Show it. OK to use the word "novel" in the strictly technical sense, but even then use sparingly.
+
+## Getting by with a Little Help from Your Friends
+- Homework for talking to your PO: NIH Guide, target IC website, RePORTER. Remember they are not only a go-between, but an advocate.
+- Know when your PO truly can't tell you anything - wait for your summary statement (not just percentile/score) before asking for revision advice, then notice of award
+- write so that an educated friend could get through your narrative in <1h, ideally less, certainly not more
+- might be nice to go through all these notes, quick runthrough of the book again, to pull out all the PO-related tips
+
+## Before and after Your Study Section Meets
+- Cleared Concepts are available either on dedicated websites or in the Council Minutes (might be a good target for a webscrape - are these minutes in plaintext or PDF?)
+
+## Is the Check in the Mail?
+- Proportion of applications discussed: NCI 55.6% of grant applications, 46.6% career development applications. NHLBI 57.6% grants, 65.9% career dev. Also, these two are the big dogs in terms of pure number of grants reviewed - the Appendix in this book has nice summaries of all the ICs, would be worth looking into other sources for funding for the type of research I'm into: data science, healthcare disparities, etc.
+- PO is the one who advocates for select pay and pay by exception (see, not just a go-between!)
+- It's ok to let your PO know if you have tenure evaluation or some other time-sensitive career event coming up so they put you on speed dial, but know that there are tons of other PIs also asking for that special treatment
+- Early stage investigators can resubmit 6wk after receipt of summary statement, but make sure it's actually a good idea to do so (how much stronger would the A1 be if you had more prelim data, etc.?)
+- It's a Good Idea to start revising your A0 immediately after you turn it in. It will either help you advance scientifically (the research itself or drafting manuscripts, abstracts) or get you a head start on your A1, or both.
+
+## The Check is Not in the Mail...
+- A1 must be w/in 37mo of A0, but this is too much time. Either the field will have advanced to the point that you need a new A0 to be current, or the field is stagnant and funding is unlikely (though I would submit that there's a slim possibility that the time for the first A0 was a little too early, but you should be reworking it during that time anyway and reevaluating)
+- on long term strategy:
+ - see if your work might be of interest to multiple ICs. Example they give is HPV in the context of head and neck cancer - could involve NCI, National Institute of Dental and Craniofacial Research (NIDCR), National Institute of Allergy and Infectious Diseases (NIAID)
+ - remember the NIH isn't the only player in the game. DoD, NSF, Agency for Healthcare Research and Quality, etc., etc.
+
+## The Check is in the Mail, but...
+- Change of Institution - awards are typically tied to you, not your institution, but there is red tape and negotiation (and tact!) necessary when making a move
+- Carryover of funds: if <25% of annual budget, no approval or explanation needed
+- No Cost Extension: usually easy to get, no application, lets you carry funds in the year after the award period, can extend to 2y with approval of PO.
+
+## Appendix
+- some interesting organizations to check out, tailored to my interests (webpages in the book, links may be added here at a future date):
+ - NCI - Cancer Control and Population Sciences; Cancer Treatment and Diagnosis
+ - NHLBI - Blood Diseases and Resources
+ - NIEHS - Risk and Integrated Sciences; Susceptibility and Population Health
+ - NIGMS - Biomedical Technology, Bioinformatics, and Computational Biology
+ - National Institute of Minority Health and Healthcare Disparities
+ - NLM might be fun for NLP work, web scraping stuff, etc.
+- A related to-do: I know the researchers I follow that have R01 funding, but haven't looked deeper. Which orgs are funding the cool data science and crossover stuff for my favorite PIs?
diff --git a/2020-11-07-on-matching.md b/2020-11-07-on-matching.md
@@ -0,0 +1,24 @@
+---
+layout: post
+title: "I matched. What does that mean, and so what?"
+toc: true
+image: https://source.unsplash.com/OfMq2hIbWMQ
+tags:
+ - personal
+ - "medical training"
+ - residency
+ - fellowship
+ - "Harrison Society"
+---
+
+## Who is this post for?
+
+This post is mostly for family and friends who are curious about how physician training works, where I am in the process, and why what just happened is awesome.
+
+## Getting into medical school is the risky part
+
+I am about to finish medical school. It was hard, but the conclusion was, largely, known from the outset. By the numbers, getting *into* medical school is much more difficult than getting out of it successfully. Most people pass their exams and graduate. Put into monetary terms, the likelihood that a person will make a physician salary is astronomically high from the second they receive their acceptance email/phone call into medical school. Of course, the fact that a doctor finished medical school tells you nothing about whether or not you would like to have him or her taking care of you or your family.
+
+## Getting into the residency and fellowship *you want* is the next risky part
+
+In the olden days, most doctors began practice immediately after graduation from medical school. Over time, it became common to do an internship, or a year-ish of on-the-job training, in a particular specialty. Eventually a longer period of training called "residency" became commonplace ("residency" was named that because the doctors literally lived in the hospital, which is still not far from the truth). Many physicians start practicing independently after residency, but a large number also go on to do a fellowship, or period of subspecialty training. (For a delightful and brief history of the stages of medical training and how the match works, see Bryan Carmody's [blog series](https://thesheriffofsodium.com/2020/01/26/the-match-part-1-why-do-we-have-a-match/)).
diff --git a/2099-03-07-on-blog-tone.md b/2099-03-07-on-blog-tone.md
@@ -0,0 +1,79 @@
+---
+layout: post
+title: "On blog tone"
+toc: true
+image: https://source.unsplash.com/OfMq2hIbWMQ
+tags:
+ - carnivalesque
+ - writing
+---
+
+## What's a blog for, anyway?
+
+
+
+The main problem with the approach I outlined was how to get those nicely updated paragraphs into the document you are sharing with colleagues.
+
+Medicine, in particular, seems wed to Microsoft Word documents for manuscripts. Word does not have a great way to include text from arbitrary files, forcing the physician-scientist to manually copy and paste those beautifully automated paragraphs. As I struggled with this, I thought (here cue Raymond Hettinger), "There must be a better way."
+
+Turns out that better way exists, and it is R Markdown.
+
+Though I was at first resistant to learning about R Markdown, mostly because I am proficient in Python and thought the opportunity cost for learning R at this point would be too high, as soon as I saw it demoed I changed my tune. Here's why.
+
+## Writing text
+- R Markdown is mostly markdown.
+ - Markdown is by far the easiest way to write plaintext documents, especially if you want to apply formatting later on without worrying about the specifics while you're writing (e.g. `#` just specifies a header - you can decide how you want the headers to look later, and that styling will automatically be applied).
+ - Plaintext is beautiful. It costs nearly nothing in terms of raw storage, and is easy to keep within a version control system. Markdown plaintext is human-readable whether or not the styling has been applied. Your ideas will never be hidden in a proprietary format that requires special software to read.
+ - I had been transitioning to writing in Markdown anyway, so +1 for R Markdown.
+- R Markdown is also a little LaTeX.
+ - LaTeX is [gorgeous](https://tex.stackexchange.com/questions/1319/showcase-of-beautiful-typography-done-in-tex-friends) and wonderful, the most flexible and expressive of all the typesetting tools (though not as fast as our old friend Groff...). It also has a steeper learning curve than Markdown, and is not so pretty on the screen in its raw form. R Markdown lets you do the bulk of your work in simple Markdown, then seamlessly invoke LaTeX when you need something a little fancier.
+- R Markdown is also a little HTML.
+ - HTML is also expressive, and can be gorgeous and wonderful. It is a pain to write. As with LaTeX, you can simply drop in some HTML where you need it, and R Markdown will deal with it as necessary.
+- R Markdown is academic-friendly.
+ - Citations and formatting guidelines for different journals are the tedious banes of any academic's existence. R Markdown has robust support for adding in citations that will be properly formatted in any desired style, just by changing a tag at the top of the document. Got a rejection from Journal 1 and want to submit to Journal 2, which has a completely different set of citation styles and manuscript formatting? NBD.
+
+## Writing code
+R Markdown, as the name implies, can also run R code.
+Any analysis you can dream of in R can be included in your document, and you can choose whether you want to show the code and its output, the output alone, or the code alone.
+People will think you went through all the work of making that figure, editing it in PowerPoint, screenshotting it to a .png, then dropping that .png file into your manuscript, but the truth is...
+you scripted all of that, so the manuscript itself made the .png and included it where it needed to go.
+
+R Markdown is by no means restricted to R code.
+This is the killer app that won me over.
+Simply by specifying that a given code block is Python,
+and installing a little tool (`reticulate`) that allows R to interface with Python,
+I can run arbitrary Python code within the document and capture the output however I want.
+That results paragraph? Sure.
+Fancy images of predictions from my machine learning model? But of course.
+
+If you don't want to use any R code ever, that's fine. R Markdown doesn't mind.
+Use SAS, MATLAB (via Octave), heck, even bash scripts - the range of language support is fantastic.
+
+## Working with friends
+R Markdown can be compiled to pretty much any format you can dream of.
+My current setup simultaneously puts out an HTML document (that can be opened in any web browser), a PDF (because I love PDFs), and (AND!) a .docx Word file,
+all beautifully formatted, on demand, whenever I hit my keyboard shortcut. I can preview the PDF or HTML as I write, have a .docx to send to my PI, and life is good.
+
+Also, because you can write in any programming language, you can easily collaborate between researchers that are comfortable in different paradigms.
+You can pass data back and forth between your chosen languages (for me, R and Python),
+either directly or by saving intermediate data to a format that both languages can read.
+
+## Automating tasks
+Many analyses and their manuscripts, especially if they use similar techniques (e.g. survival modeling), are rather formulaic.
+Many researchers have scripts they keep around and tweak for new analyses revolving around the same basic subject matter or approach.
+With R Markdown, your entire manuscript becomes a runnable program, further automating the boring parts of getting research out into the open.
+
+One of the [first introductions](https://www.youtube.com/watch?v=MIlzQpXlJNk) I had to R Markdown shared the remarkable idea of setting the file to run on a regular basis,
+generating a report based on any updated data,
+and then sending this report to all the interested parties automatically.
+While much academic work could not be so fully automated, parts of it certainly can be.
+
+Perhaps your team is building a database for outcomes in a given disease, and has specified the analysis in great detail beforehand.
+One of my mentors gives the advice that in any project proposal you should go as far as to mock up the results section,
+including all figures,
+so you make sure you are collecting the right data.
+If this was done in an R Markdown document rather than a simple Word document,
+you could have large parts of the template manuscript
+become the real manuscript as the database fleshes out over time.
+Then when it's done, look over the data, make additions and subtractions as needed,
+write the discussion sections, and send it in.
diff --git a/2099-04-13-wget-outta-my-way.md b/2099-04-13-wget-outta-my-way.md
@@ -0,0 +1,35 @@
+---
+layout: post
+title: "Wget Outta My Way, diigo"
+toc: true
+image: https://source.unsplash.com/OfMq2hIbWMQ
+tags:
+ - wget
+ - web
+ - archiving
+ - productivity
+ - Python
+ - Markdown
+ - "academic writing"
+---
+
+## read-later
+
+I've been thinking a lot about sustainable, preferably third-party-service-free ways to keep track of and use things I've read online.
+
+[wget manual online](https://www.gnu.org/software/wget/manual/wget.html#Download-Options)
+
+`wget -E -k -p https://www.nateliason.com/blog/smart-notes`
+
+- `-E` - adds the `.html` extension to the filename
+- `-p` - page requisites (downloads all resources necessary to properly render the page, e.g. images)
+- `-nd` - no directories
+- `-nH` - no host directories
+- `-H` - spans hosts when recursively retrieving
+- `-K` - "When converting a file, back up the original version with a ‘.orig’ suffix."
+- `-k` - convert non-relative links
+- `-P` - specify directory for download, creates if it doesn't exist. Append desired directory name directly to the command, e.g. a directory called "web" would be specified as `-Pweb`
+
+"Actually, to download a single page and all its requisites (even if they exist on separate websites), and make sure the lot displays properly locally, this author likes to use a few options in addition to ‘-p’:"
+
+`wget -E -H -k -K -p -Pweb https://www.nateliason.com/blog/smart-notes`
diff --git a/2099-06-10-on-old-software.md b/2099-06-10-on-old-software.md
@@ -0,0 +1,43 @@
+---
+layout: post
+title: "On Old Software"
+toc: false
+image: https://en.wikipedia.org/wiki/Vi#/media/File:Vi_source_code_join_line_logic.png
+categories:
+ - AI for MDs
+tags:
+ - coding
+ - software
+ - history
+ - laziness
+ - groff
+ - latex
+ - vim
+ - gopher
+---
+
+ a later date.[^4]
+
+# Typesetting
+
+# Text editing
+
+# Internet
+
+
+[^6]: You may have noticed that the name of this file is "results_paragraphs_latex.txt" rather than "results_paragraphs.txt," and that's because LaTeX needs a little special treatment if you're going to use the percentage symbol. LaTeX uses the percentage symbol as a comment sign, meaning that anything after the symbol is ignored and left out of the document. You have to "escape" the percentage symbol with a slash, like this: `\%`. I have this simple bit of code that converts the normal text file into a LaTeX-friendly version:
+
+ ```python
+ # make a LaTeX-friendly version (escape the % symbols with \)
+ # Read in the file
+ with open(results_text_file, "r") as file:
+ filedata = file.read()
+ # Replace the target string
+ filedata = filedata.replace("%", "\%")
+ # Write the file
+ text_file_latex = "results_paragraphs_latex.txt"
+ with open(text_file_latex, "w") as file:
+ file.write(filedata)
+ ```
+
+[^7]: You may have noticed there are two datasets I'm pulling from for this, "data," which includes everything on the basis of _hospitalizations_, and "df," short for "dataframe," which is a subset of "data" that only includes each _patient_ once (rather than a new entry for every hospitalization), along with a few other alterations that allow me to do patient-wise calculations.
diff --git a/2099-06-10-on-video-games.md b/2099-06-10-on-video-games.md
@@ -0,0 +1,26 @@
+---
+layout: post
+title: "On Old Software"
+toc: false
+image: https://en.wikipedia.org/wiki/Vi#/media/File:Vi_source_code_join_line_logic.png
+categories:
+ - AI for MDs
+tags:
+ - coding
+ - software
+ - history
+ - laziness
+ - groff
+ - latex
+ - vim
+---
+
+# Video games
+http://www.cnn.com/2010/TECH/gaming.gadgets/08/18/video.game.history/index.html
+https://www.theatlantic.com/technology/archive/2016/04/how-early-computer-games-influenced-internet-culture/478167/
+https://en.wikipedia.org/wiki/History_of_video_games
+https://www.jesperjuul.net/thesis/2-historyofthecomputergame.html
+
+[^5]: Assign it to a resident, of course.
+
+[^7]: You may have noticed there are two datasets I'm pulling from for this, "data," which includes everything on the basis of _hospitalizations_, and "df," short for "dataframe," which is a subset of "data" that only includes each _patient_ once (rather than a new entry for every hospitalization), along with a few other alterations that allow me to do patient-wise calculations.
diff --git a/2099-06-11-lightgbm-multiclass.md b/2099-06-11-lightgbm-multiclass.md
@@ -0,0 +1,132 @@
+---
+layout: post
+title: "Python, Write My Paper"
+toc: false
+image: /images/unsplash-grey-flowerbuds.jpg
+categories:
+ - AI for MDs
+tags:
+ - coding
+ - python
+ - fstring
+ - laziness
+---
+
+{:class="img-responsive"}
+
+Computers are good at doing tedious things.[^1]
+
+Many of the early advances in computing were accomplished to help people do tedious things they didn't want to do, like the million tiny equations that make up a calculus problem.[^2] It has also been said, and repeated, and I agree, that one of the three virtues of a good programmer is laziness.[^3]
+
+One of the most tedious parts of my job is writing paragraphs containing the results of lots of math relating to some biomedical research project. To make this way easier, I use a core Python utility called the `f-string`, in addition to some other tools I may write about at a later date.[^4]
+
+## The problem
+
+First, here's an example of the kinds of sentences that are tedious to type out, error prone, and have to be fixed every time something changes on the back end (--> more tedium, more room for errors).
+
+"In the study period there were 1,485,880 hospitalizations for 708,089 unique patients, 439,696 (62%) of whom had only one hospitalization recorded.
+The median number of hospitalizations per patient was 1 (range 1-176, [1.0 , 2.0])."
+
+The first paragraph of a results section of a typical medical paper is chock-full of this stuff. If we find an error in how we calculated any of this, or find that there was a mistake in the database that needs fixing (and this happens woefully often), all of the numbers need replaced. It's a pain.
+How might we automate the writing of this paragraph?[^5]
+
+## The solution
+
+First, we're going to do the math (which we were doing anyway), and assign each math-y bit a unique name. Then we're going to plug in the results of these calculations to our sentences.
+If you're not familiar with Python or Pandas, don't worry - just walk through the names and glance at the stuff after the equals sign, but don't get hung up on it.
+The basic syntax is:
+
+```python
+some_descriptive_name = some_dataset["some_column_in_that_dataset"].some_mathy_bit()
+```
+
+After we generate the numbers we want, we write the sentence, insert the code, and then use some tricks to get the numbers in the format we want.
+
+In most programming languages, "string" means "text, not code or numbers." So an `f-string` is a `formatted-string`, and allows us to insert code into blocks of normal words using an easy, intuitive syntax.
+
+Here's an example:
+
+```python
+name_of_word_block = f"""Some words with some {code} we want Python to evaluate,
+maybe with some extra formatting thrown in for fun,
+such as commas to make long numbers more readable ({long_number:,}),
+or a number of decimal places to round to
+({number_with_stuff_after_the_decimal_but_we_only_want_two_places:.2f},
+or a conversion from a decimal to a percentage and get rid of everything after the '.'
+{some_number_divided_by/some_other_number*100:.0f}%)."""
+```
+
+First, declare the name of the block of words. Then write an `f`, which will tell Python we want it to insert the results of some code into the following string, which we start and end with single or triple quotes (triple quotes let you break strings into multiple lines).
+Add in the code within curly brackets, `{code}`, add some optional formatting after a colon, `{code:formatting_options}`, and prosper.
+
+As you can see from the last clause, you can do additional math or any operation you want within the `{code}` block. I typically like to do the math outside of the strings to keep them cleaner looking, but for simple stuff it can be nice to just throw the math in the f-string itself.
+
+Here's the actual code I used to make those first two sentences from earlier. First the example again, then the math, then the f-strings.[^7]
+
+"In the study period there were 1,485,880 hospitalizations for 708,089 unique patients, 439,696 (62%) of whom had only one hospitalization recorded.
+The median number of hospitalizations per patient was 1 (range 1-176, [1.0 , 2.0])."
+
+```python
+n_encs = data["encounterid"].nunique()
+n_pts = data["patientid"].nunique()
+
+pts_one_encounter = df[df["encounternum"] == 1].nunique()
+min_enc_per_pt = df["encounternum"].min()
+q1_enc_per_pt = df["encounternum"].quantile(0.25)
+median_enc_per_pt = df["encounternum"].median()
+q3_enc_per_pt = df["encounternum"].quantile(0.75)
+max_enc_per_pt = df["encounternum"].max()
+
+sentence01 = f"In the study period there were {n_encs:,} hospitalizations for {n_pts:,} unique patients, {pts_one_encounter:,} ({pts_one_encounter/n_pts*100:.0f}%) of whom had only one hospitalization recorded. "
+sentence02 = f"The median number of hospitalizations per patient was {median_enc_per_pt:.0f} (range {min_enc_per_pt:.0f}-{max_enc_per_pt:.0f}, [{q1_enc_per_pt} , {q3_enc_per_pt}]). "
+```
+
+If you want to get real ~~lazy~~ ~~fancy~~ lazy, you can combine these sentences into a paragraph, save that paragraph to a text file, and then automatically include this text file in your final document.
+
+```python
+paragraph01 = sentence01 + sentence02
+results_text_file = "results_paragraphs.txt"
+with open(results_text_file, "w") as text_file:
+ print(paragraph01, file=text_file)
+```
+
+To automatically include the text file in your document, you'll have to figure out some tool appropriate to your writing environment. I think there's a way to source text files in Microsoft Word, though I'm less familiar with Word than other document preparation tools such as LaTeX. If you know how to do it in Word, let me know (or I'll look into it and update this post).
+
+Here's how to do it in LaTeX. Just put `\input` and the path to your text file at the appropriate place in your document:[^6]
+
+```latex
+\input{"results_paragraphs_latex.txt"}
+```
+
+With this workflow, I can run the entire analysis, have all the mathy bits translated into paragraphs that include the right numbers, and have those paragraphs inserted into my text in the right spots.
+
+I should note that there are other ways to do this. There are ways of weaving actual Python and R code into LaTeX documents, and RMarkdown is a cool way of using the simple syntax of Markdown with input from R. I like the modular approach outlined here, as it lets me just tag on a bit to the end of the Python code I was writing anyway, and integrate it into the LaTeX I was writing anyway. I plan to use this approach for the foreseeable future, but if you have strong arguments for why I should switch to another method, I would love to hear it, especially if it might better suit my laziness.
+
+Addendum: As I was writing this, I found a similar treatment of the same subject. It's great, with examples in R and Python. [Check it out](https://jabranham.com/blog/2018/05/reporting-statistics-in-latex/).
+
+[^1]: _Automate the Boring Stuff_ by Al Sweigart is a great introduction to programming in general, and is available for free as a [hypertext book](https://automatetheboringstuff.com/). It teaches exactly what its name denotes, in an interactive and easy-to-understand combination of code and explanation.
+
+[^2]: I'm revisiting [Walter Isaacson's _The Innovators"](https://en.wikipedia.org/wiki/The_Innovators_(book)), which I first listened to before I got deeply into programming, and on this go-through I am vibing much harder with the repeated (and repeated) (and again repeated) impetus for building the first and subsequent computing machines: tedious things are tedious.
+
+[^3]: The other two are impatience and hubris. Here is one of the [most lovely websites on the internet](http://threevirtues.com/)
+
+[^4]: For example, TableOne, which makes the (_incredibly_ tedious) task of making that classic first table in any biomedical research paper _so much easier_. Here's a link to [TableOne's project page](https://github.com/tompollard/tableone), which also includes links out to examples and their academic paper on the software.
+
+[^5]: Assign it to a resident, of course.
+
+[^6]: You may have noticed that the name of this file is "results_paragraphs_latex.txt" rather than "results_paragraphs.txt," and that's because LaTeX needs a little special treatment if you're going to use the percentage symbol. LaTeX uses the percentage symbol as a comment sign, meaning that anything after the symbol is ignored and left out of the document. You have to "escape" the percentage symbol with a slash, like this: `\%`. I have this simple bit of code that converts the normal text file into a LaTeX-friendly version:
+
+ ```python
+ # make a LaTeX-friendly version (escape the % symbols with \)
+ # Read in the file
+ with open(results_text_file, "r") as file:
+ filedata = file.read()
+ # Replace the target string
+ filedata = filedata.replace("%", "\%")
+ # Write the file
+ text_file_latex = "results_paragraphs_latex.txt"
+ with open(text_file_latex, "w") as file:
+ file.write(filedata)
+ ```
+
+[^7]: You may have noticed there are two datasets I'm pulling from for this, "data," which includes everything on the basis of _hospitalizations_, and "df," short for "dataframe," which is a subset of "data" that only includes each _patient_ once (rather than a new entry for every hospitalization), along with a few other alterations that allow me to do patient-wise calculations.
diff --git a/2099-12-31-coding_isnt_scary_its_awesome.md b/2099-12-31-coding_isnt_scary_its_awesome.md
@@ -0,0 +1,31 @@
+---
+layout: post
+title: "Coding is awesome and also not scary"
+categories:
+ - technical
+tags:
+ - machine learning
+ - programming
+ - education
+ - meta
+---
+
+# Coding isn't scary. It's awesome.
+
+I want to show you something.
+
+It's a tiny piece of computer code. And it's freaking rad.
+
+Ready?
+
+`AllTheData.describe`
+
+Looks simple enough. All I did was enter the name of my dataset, `AllTheData`, add a period, and write the word `describe`. Then I hit the play button and waited 0.143 milliseconds. And then I saw...
+
+# (output from some data set)
+
+Yup. Averages, medians, and quartiles for every feature in my dataset. Table 1, if you will. With a single command, which I can type faster than I can go clicking around Excel.
+
+This is why computers are cool, and why they're even cooler when we control them with code. We can do very useful but very complicated things with simple commands. Under the surface, that `.describe` command does a whole bunch of math that would be a pain to do by hand, and because we do this type of math so much someone made it dead simple to execute.
+
+Let me show you one more example. It's essentially the same
diff --git a/2099-12-31-goals-for-this-blog.md b/2099-12-31-goals-for-this-blog.md
@@ -0,0 +1,29 @@
+---
+layout: post
+title: "Goals for this blog"
+categories:
+ - medicine
+tags:
+ - machine learning
+ - programming
+ - education
+ - meta
+---
+
+# Goals
+
+
+# Post Ideas
+
+- All the tables and figures for your paper in X lines of code (table 1, uni/multivariate analyses, cox survival, kaplan meier, etc.) (maybe use this article as a guide: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0077301 . Table 3 has list of top statistical measures and methods)
+- Clean your data in 1 line of code (most common data-entry problems)
+- Pick the best machine learning model for your data with one chart
+- Maguru panggul for code
+-
+Why should physicians learn to code?
+ - Not every physician needs to learn to code. We already have too much to learn and retain. Administrative work eats up our time outside of (or, too often, while) seeing patients. Even for those with "protected" research time, it is consumed by managing the lab, directing resources, and catching up on patient notes from yesterday, while actual scientific writing happens in time carved out of sleep in the late night or early morning. And what about our loved ones and the occasional fit of self-care? In other words, we are physician-scientists too, and we get it. It's hard. However, our goal in this book is to show how a little knowledge of basic programming principles, combined with practical examples you can copy, paste, and modify for your own use, can save time, enable analyses that were previously unthinkable, improve communication, help you grasp (and critique) the latest research, and, ultimately, do what we all came here to do: help people, using all of the scientific tools and human compassion we can muster.
+ - Another argument against learning to code is the proliferation of point-and-click interfaces for doing machine learning, as well as companies that sell their programming services for modest fees. If other people have done or will do the coding for you, why learn? We have nothing against this approach, and hope that machine learning tools will continue to become more accessible while specialist companies become profitable. There is room for everyone. Three arguments for learning to do it yourself are that you can remain free, open-source, and up-to-date.
+ - Free: Python and R are free programming languages. The algorithms you need for the vast majority of machine learning applications are prepackaged in free libraries. Proprietary and expensive software may offer niche advantages in particular content areas, but this is less and less true as the years go on and people publish equivalent yet free solutions. Companies and specialists that do machine learning for you are great, and can help with your most complex problems, but most of the time, frankly, you will have a bread-and-butter dataset you can analyze with simple code on your own computer. If you do it yourself you won't have to deal with financial, legal, IRB, and HIPAA beyond what you did to get the project going in the first place. You also have irreplaceable content expertise: you know your patients, the medicine, what questions to ask, and what bogus results look like. Finally, maybe your machine learning idea _won't_ work, but you won't know that until you've run a quick pilot. When you do work with a specialist, how much more powerful and streamlined will your conversation be if you have pilot data, or at least specific problems, to discuss? With a modest computer (most of our analyses are done on a low-powered hospital-supplied tablet PC) and a few free downloads, you can start today, no poorer or more frustrated with paperwork than when you started. This also means that researchers around the world, regardless of financial circumstance, can make an impact. Lastly, if you are an educator, there are no financial barriers to introducing your students to these concepts and tools. They're _free_.
+ - Open-source 1: Python and R are open-source. Not only do you not have to pay for them, but you can crack them open and interrogate them all the way down to the 1s and 0s. Nothing is proprietary or hidden. You can modify them to suit your needs, or even publish your own custom version. This is also true for the majority of machine learning packages. Scientific reproducibility is at its maximum when the tools used for analyses are completely transparent. It is becoming common for researchers to publish not only their (HIPAA-compliant) raw data, but also the actual code. "Go ahead, run it and see if you get what we got." If you write the code yourself, you can easily share it, receive critique, and improve it. This is good science.
+ - Open-source 2: When you see the words "open-source," you should immediately think "community." Researchers publish code not only to officially claim ideas, but to engage with communities. Online repositories such as GitHub make it easy to ask questions, suggest improvements, and give examples. We continue to be impressed by how willing researchers are to participate in community, and have had many productive conversations with authors and fellow users, including requests for features (that were then implemented), questions about problems, and suggestions about uses. People are excited for you to use their software, and want to help you take it in new directions. There are other programming-specific communities such as StackExchange, where programmers in every different field present their coding problems, some quite granular, and others give suggestions that are dynamically peer-reviewed and preserved online. The best answers have code you can copy and paste, and there are typically several approaches offered for the same problem. One of the first steps to becoming a good programmer is learning how to Google for your problem, find an answer (usually on GitHub or Stack), and modify the code you copied and pasted. You almost never have to reinvent any wheels. You just change out hubcaps and bolts here and there. This is possible because of the robust and generous communities that have built up around open-source software and ideals.
+ - Up-to-date: The state of the art is online. When machine learning scientists publish new algorithms, they often publish an R or Python package to go along with the paper, so you can try it out right away. It takes time for these algorithms to make it into point-and-click interfaces, and some algorithms may never be available as an option in a prebuilt system. When they do show up, it may not be able to perform the necessary tweaks and adjustments to fit your use. (Imagine if you could only prescribe combination pills, and had no way to adjust the dosage of each component!) A little programming experience gives you the power and flexibility to use algorithms, new and old, in any combination you see fit. You will find that, compared to point-and-click interfaces, most tasks are easier and faster once you have the code for them, and you can always update them to suit your needs and preferences.
diff --git a/2099-12-31-on-the-logo-and-its-color.md b/2099-12-31-on-the-logo-and-its-color.md
@@ -0,0 +1,30 @@
+---
+layout: post
+title: "On the logo and its color"
+categories:
+ - medicine
+tags:
+ - arsenic
+ - cancer
+ - iconography
+ - color
+ - medicine
+---
+
+## Arsenic
+
+
+
+On my first day in leukemia clinic in 2015, with a [certain young fellow from Syria](https://my.clevelandclinic.org/staff/20358-aziz-nazha) and a [certain attending physician who is rather fond of a good story](https://www.nytimes.com/by/mikkael-a-sekeres), I was told the story of how arsenic made its way back into medicine as a treatment (and often cure) for one of the worst types of blood cancer, "Acute Promyelocytic Leukemia" (APL).
+
+Arsenic has been used for [thousands of years](https://doi.org/10.1007/s00204-012-0866-7) for everything from [medicine to candy to wallpaper to rat/king poison](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1036521/). It was used widely for this last purpose, because it is [odorless, tasteless, dissolves instantly in liquid, and is among the more deadly poisons known to man](https://giphy.com/explore/the-princess-bride). In low doses it is a mild stimulant (and, apparently, aphrodisiac), and in higher doses bad things happen, [even if not ingested](https://wellcomecollection.org/works/g32gywph).
+
+In the 1800s in the West arsenic was found to be helpful in certain blood cancers if ingested and in certain skin and breast cancers if applied to the skin. A string of studies from China in the 1990s using arsenic trioxide for patients with APL had remarkable results, which led to studies in the U.S and a remarkably swift change in standard medical practice. APL was a disease that would almost invariably bring you into the hospital in critical condition and send you out in a bag to. Now, last time I was with a patient with APL, that young Syrian fellow (who is now an attending) kept repeating:
+
+↵ Sun HD, Ma L, Hu X-C et al. Ai-Lin 1 treated 32 cases of acute promyelocytic leukemia. Chin J Integrat Chin Western Med 1992;12:170-172. Google Scholar
+Zhang P, Wang SY, Hu XH. Arsenic trioxide treated 72 cases of acute promyelocytic leukemia. Chin J Hematol 1996;17:58-62. Google Scholar
+↵ Shen Z-X, Chen G-Q, Ni J-H et al. Use of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia (APL): II. Clinical efficacy and pharmacokinetics in relapsed patients. Blood 1997;89:3354-3360. Abstract/FREE Full Text
+↵ Soignet SL, Maslak P, Wang Z-G et al. Complete remission after treatment of acute promyelocytic leukemia with arsenic trioxide. N Engl J Med 1998;339:1341-1348. CrossRefMedlineGoogle Scholar
+↵ Soignet S, Frankel S, Tallman M et al. U.S. multicenter trial of arsenic trioxide (AT) in acute promyelocytic leukemia (APL) [abstract 3084]. Blood 1999;94(suppl 10):698a. Google Scholar
+
+[](https://wellcomecollection.org/works/z8ssmfqa?query=arsenic&page=1)
diff --git a/2099-12-31-zotero.md b/2099-12-31-zotero.md
@@ -0,0 +1,66 @@
+---
+layout: post
+title: "On the pretentious Latin slogan"
+categories:
+ - medicine
+tags:
+ - arsenic
+ - cancer
+ - balance
+ - humility
+ - iconography
+ - color
+---
+
+# sola dosis facit venenum
+
+### Everything is a poison, everything is a medicine.
+
+First, two quotes, one in English and one in German.
+
+Don't worry, we'll translate the German. This isn't one of those 19th century novels that expects you to know English, French, Latin, Italian, and German just to get through a page. I left the quote in the original because one of the words has a delightful dual meaning that is lost if not juxtaposed with an English translation.
+
+> Poisons in small doses are the best medicines; and the best medicines in too large doses are poisonous.
+
+> <cite><a href="http://theoncologist.alphamedpress.org/content/6/suppl_2/1.long">William Withering, 18th century* English physician, discoverer of digitalis (sort of), and proponent of arsenic therapy.</a></cite>
+
+>Alle Dinge sind Gift, und nichts ist ohne Gift; allein die dosis machts, daß ein Ding kein Gift sei.
+
+> <cite><a href="http://www.zeno.org/Philosophie/M/Paracelsus/Septem+Defensiones/Die+dritte+Defension+wegen+des+Schreibens+der+neuen+Rezepte">From the 1538 *Septum Defensiones* by Philippus Aureolus Theophrastus Bombastus von Hohenheim, later known as Paracelsus, Swiss physician, "Father of Toxicology."</a></cite>
+
+The phrase "sola dosis facit venenum," (usually rendered "the dose makes the poison" in English) is a Latinization of the German phrase above from Paracelsus. He wrote this in his "Seven Defenses" when he was fighting against accusations of poisoning his patients (malpractice court, it seems, is one of the oldest traditions in medicine).
+
+Paracelsus is the Latin name given to von Hohenheim by his friends, who were probably screwing with him, or his scribe, whose poor fingers grew to hate the eighteen syllable German name.
+
+(I'm convinced that translating a name or phrase into Latin was the historic equivalent of becoming a friend on social media: OK, now it's official.)
+
+The basic idea of the poison-medicine continuum is given plainly by Withering, but here's my rough translation from Paracelsus' German:
+
+> All things are poison, and nothing is not a poison. Only the dose makes the thing not a poison.
+
+Throw in a couple of exclamation marks, italics, and fist pounds, and you have the makings of dialogue for a 16th century *Law and Order*.
+
+It makes me happy that the word for "poison" in German is "gift." English "gift," meaning "present," and German "gift" come from the same root, "[to give](https://www.etymonline.com/word/gift)," but language is also susceptible to [divergent evolution](https://en.wikipedia.org/wiki/Divergent_evolution), sometimes [amusingly](http://www.bbc.co.uk/languages/yoursay/false_friends/german/be_careful__its_a_gift_englishgerman.shtml). This dual meaning fits nicely with the general conceit --- medicine and poison are two sides of the same coin, gifts in either case, to be used [with judgment, not to excess](https://www.lds.org/scriptures/dc-testament/dc/59.20).
+
+
+## Arsenic
+
+
+
+On my first day in leukemia clinic in 2015, with a [certain young fellow from Syria](https://my.clevelandclinic.org/staff/20358-aziz-nazha) and a [certain attending physician who is rather fond of a good story](https://www.nytimes.com/by/mikkael-a-sekeres), I was told the story of how arsenic made its way back into medicine as a treatment (and often cure) for one of the worst types of blood cancer, "Acute Promyelocytic Leukemia" (APL).
+
+Arsenic has been used for [thousands of years](https://doi.org/10.1007/s00204-012-0866-7) for everything from [medicine to candy to wallpaper to rat/king poison](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1036521/). It was used widely for this last purpose, because it is [odorless, tasteless, dissolves instantly in liquid, and is among the more deadly poisons known to man](https://giphy.com/explore/the-princess-bride). In low doses it is a mild stimulant (and, apparently, aphrodisiac), and in higher doses bad things happen, [even if not ingested](https://wellcomecollection.org/works/g32gywph).
+
+In the 1800s in the West arsenic was found to be helpful in certain blood cancers if ingested and in certain skin and breast cancers if applied to the skin. A string of studies from China in the 1990s using arsenic trioxide for patients with APL had remarkable results, which led to studies in the U.S and a remarkably swift change in standard medical practice. APL was a disease that would almost invariably bring you into the hospital in critical condition and send you out in a bag to. Now, last time I was with a patient with APL, that young Syrian fellow (who is now an attending) kept repeating:
+
+↵ Sun HD, Ma L, Hu X-C et al. Ai-Lin 1 treated 32 cases of acute promyelocytic leukemia. Chin J Integrat Chin Western Med 1992;12:170-172. Google Scholar
+Zhang P, Wang SY, Hu XH. Arsenic trioxide treated 72 cases of acute promyelocytic leukemia. Chin J Hematol 1996;17:58-62. Google Scholar
+↵ Shen Z-X, Chen G-Q, Ni J-H et al. Use of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia (APL): II. Clinical efficacy and pharmacokinetics in relapsed patients. Blood 1997;89:3354-3360. Abstract/FREE Full Text
+↵ Soignet SL, Maslak P, Wang Z-G et al. Complete remission after treatment of acute promyelocytic leukemia with arsenic trioxide. N Engl J Med 1998;339:1341-1348. CrossRefMedlineGoogle Scholar
+↵ Soignet S, Frankel S, Tallman M et al. U.S. multicenter trial of arsenic trioxide (AT) in acute promyelocytic leukemia (APL) [abstract 3084]. Blood 1999;94(suppl 10):698a. Google Scholar
+
+[](https://wellcomecollection.org/works/z8ssmfqa?query=arsenic&page=1)
+
+
+\*The article from The Oncologist that gave the Withering quote made the unfortunate mistake of saying he was a *15th* century physician, which has, even more unfortunately, led to the persistence of the wrong century attached to his name. He was born in 1741, discovered digitalis in 1775, and died in 1799.
+
diff --git a/2999-12-31-template.md b/2999-12-31-template.md
@@ -0,0 +1,24 @@
+---
+layout: post
+title: "title"
+toc: true
+image: https://source.unsplash.com/OfMq2hIbWMQ
+categories:
+ - AI for MDs
+tags:
+ - coding
+ - software
+ - laziness
+ - "R Markdown"
+ - Python
+ - Markdown
+ - "academic writing"
+---
+
+## animal
+
+Link to [local page]({% post_url 2019-06-10-python-write-my-paper %}) .
+
+
+- LaTeX is [gorgeous](https://tex.stackexchange.com/questions/1319/showcase-of-beautiful-typography-done-in-tex-friends)
+- One of the [first introductions](https://www.youtube.com/watch?v=MIlzQpXlJNk)